![]()
click on the images to jump to some results.
Electron microscopy (EM) is key to mapping the morphology of neural structures. Recent techniques, such as Focused Ion Beam Scanning Electron Microscopy (FIB-SEM), can now deliver image stacks at the nanometer resolution in all three dimensions. Such stacks show very fine structures that are critical to unlocking new insights into brain function but are still mostly analyzed by hand, which can require months of tedious labor. As a result, the vast majority of this very high quality data goes unused. Furthermore, although they contain tens of millions of voxels, these stacks span volumes smaller than 10x10x10 μm3, which presents less than a billionth of the volume of the entire brain. If it is ever to be mapped in its entirety, automation will be required. Our goal is to propose a fully automated approach that can segment large EM datasets by using sophisticated cues that capture global shape and texture.
A. Lucchi, K. Smith, R. Achanta, V. Lepetit, G. Knott, P. Fua
Results
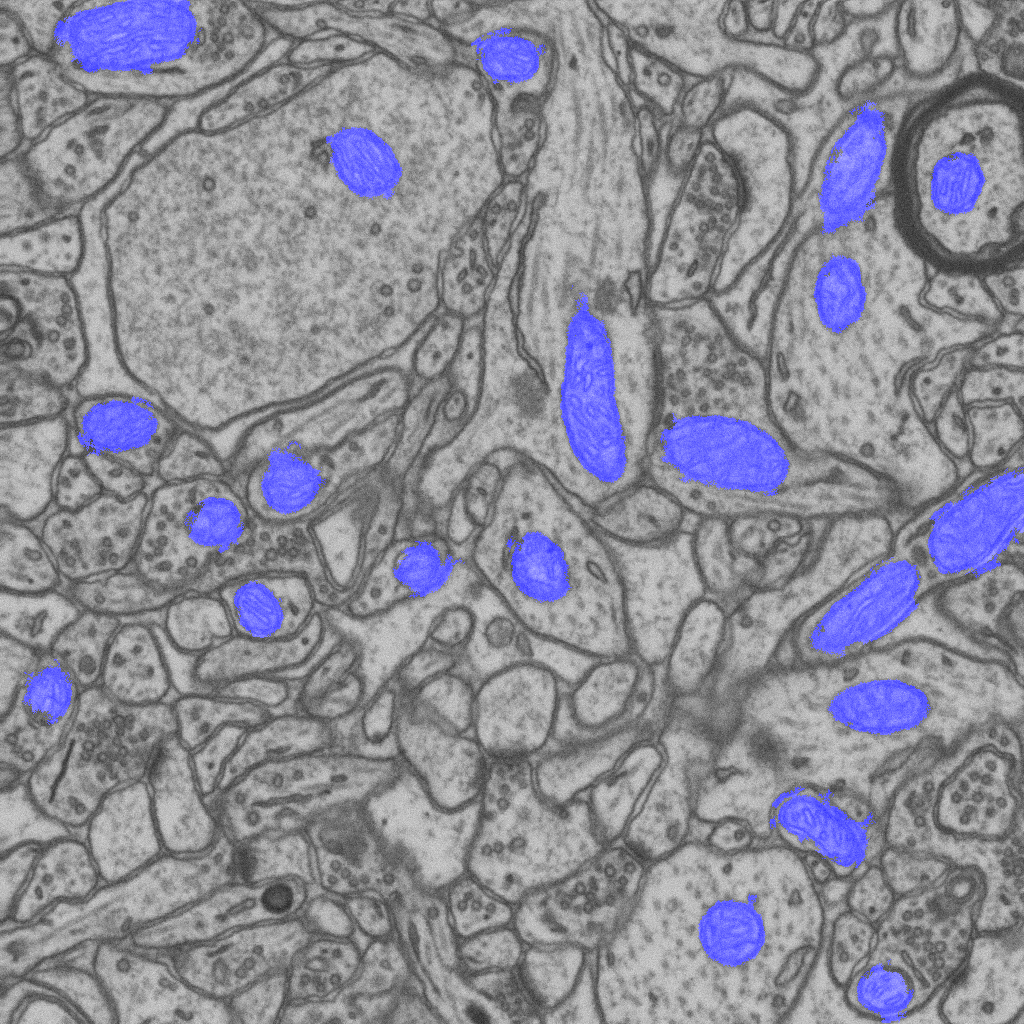
Automatic segmentation of mitochondria also called “cellular power plants” from a FIB-SEM image stack. The blue overlay indicates areas segmented as mitochondria by grouping individual voxels into supervoxels, computing shape and intensity features from these supervoxels, and feeding them to a graph-cut algorithm.
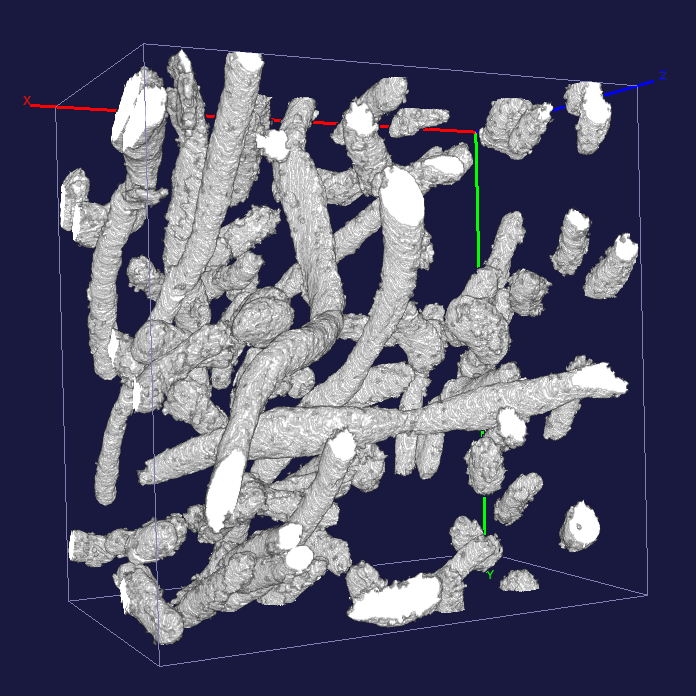
The voxels that were painted blue in the above example are carved out of the volume to produce a 3D reconstruction of the mitochondria. Note how elongated some of them are.
Materials
- Poster
- SLIC Superpixel and Supervoxel code
- C++ code for the 3d ray features available on demand
References
Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
Learning Structured Models for Segmentation of 2D and 3D Imagery
IEEE Transactions on Medical Imaging. 2015. Vol. 34, num. 5, p. 1096-1110. DOI : 10.1109/TMI.2014.2376274.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
Exploiting Enclosing Membranes and Contextual Cues for Mitochondria Segmentation
2014. International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Boston, Massachusetts, USA, September 2014.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
Learning for Structured Prediction Using Approximate Subgradient Descent with Working Sets
2013. Conference on Computer Vision and Pattern Recognition (CVPR), Portland, Oregon, USA, June 23-28, 2013. DOI : 10.1109/Cvpr.2013.259.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.
Structured Image Segmentation using Kernelized Features
2012. European Conference on Computer Vision, Florence, Italy, October 2012.Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.