Scientists in disciplines like biology, chemistry, physics etc. produce vast amounts of data through experimentation and simulation. The amounts of data produced are already so big that they can barely be managed. And the problem is certain to get worse as the volume of scientific data doubles every year. In the DIAS laboratory we are working on next generation data management tools and techniques able to manage tomorrow’s scientific data.
We study large and demanding scientific databases and are particularly interested in:
- aiding scientists with “systems” work, such as database schema, physical design, and data placement on disks (and automating all related procedures to minimize the need for human intervention)
- designing and developing computational support for popular scientific data structures (and especially ones not currently supported by cutting-edge database technology, such as tetrahedral meshes or protein structures)
- understanding and aiding the logical interpretation of data (including data cleaning, validation, and schema mapping)
The Human Brain Project
In the context of this project we address the particular problems of neuroscientists on their quest to understand and simulate the human brain. More specifically, we work together with neuroscientists in the Human Brain Project (https://www.humanbrainproject.eu) to manage the vast amounts of data they produce. Their research, modeling and simulating only fractions of the human brain, already produces terabytes of data. With the recent upgrade of computing infrastructure (IBM Blue Gene/P), the volume of data will soon be in the order of petabytes.
In order to better understand how the brain works, the neuroscientists build biophysically realistic models of a neocortical column on the molecular level and simulate them on a supercomputer. Each of the models contains several thousand neurons where each neuron and its branches are modeled as thousands of cylinders. One problem, for example, that we are continuously working on is improving neurons retrieval based on their position in three-dimensional space. This neuroscience problem translates into the data management problem of spatial data (range/distance/k-nearest neighbor) querying. Another example includes computing locations where neurons “touch” each other and form synapses. This problem translates into spatial join. While most existing (spatial indexing) techniques perform well with several million of neurons (each consisting of several thousand cylinders), soon we need to scale to billions of neurons. As the models just keep getting bigger and more detailed, the work is continuous and we are constantly investigating new techniques to address these and similar problems.
|
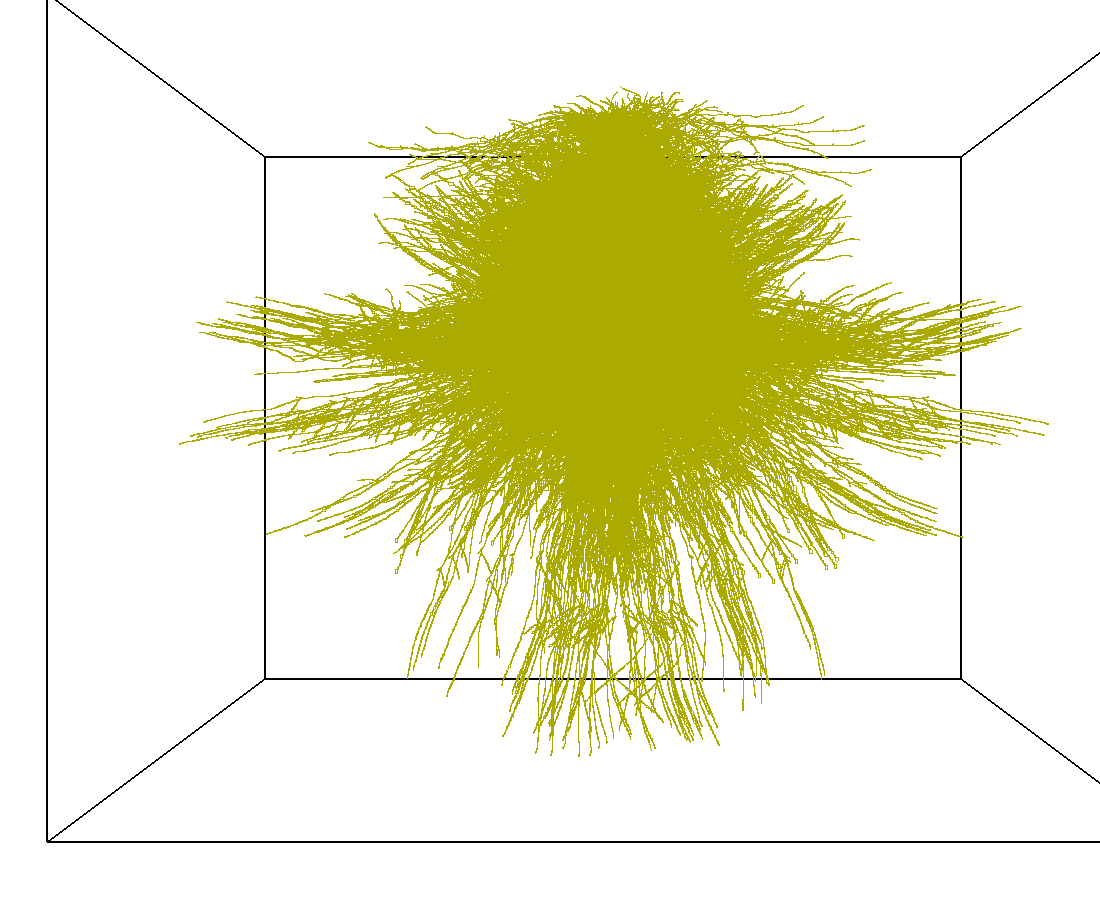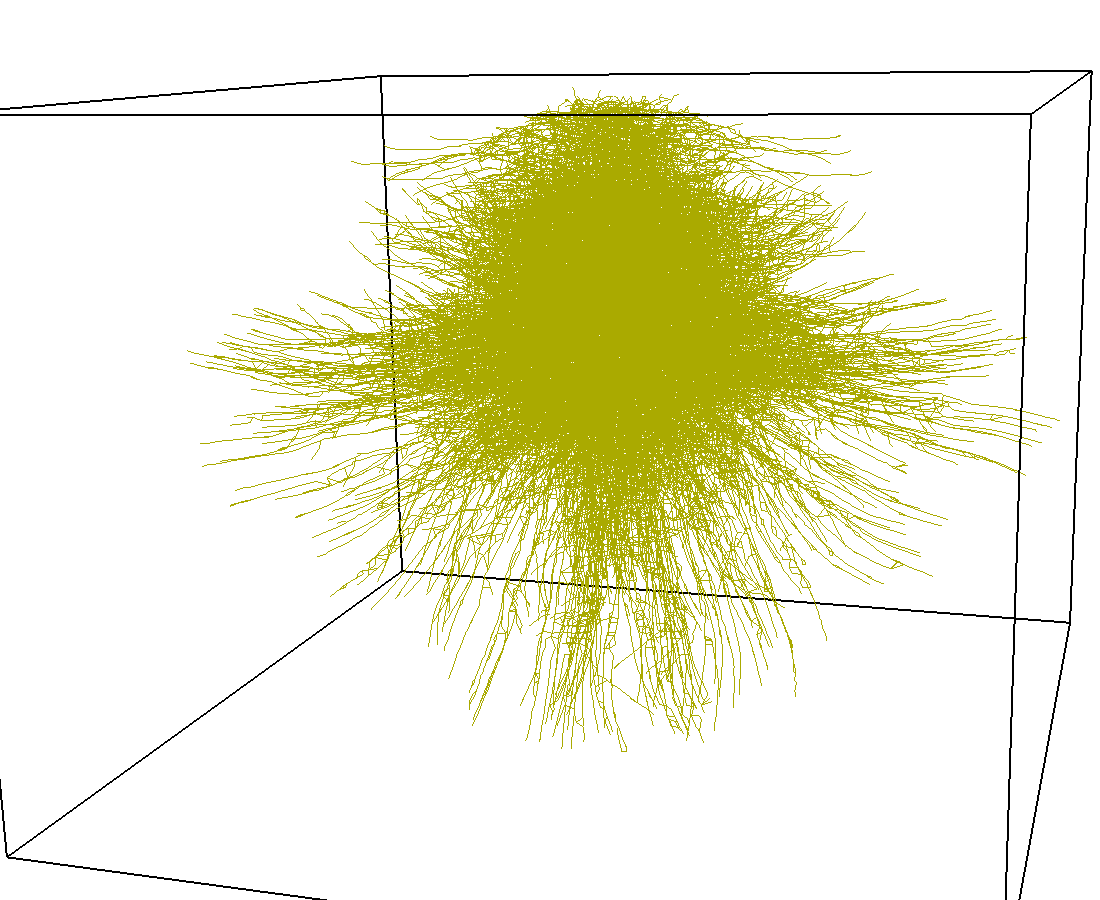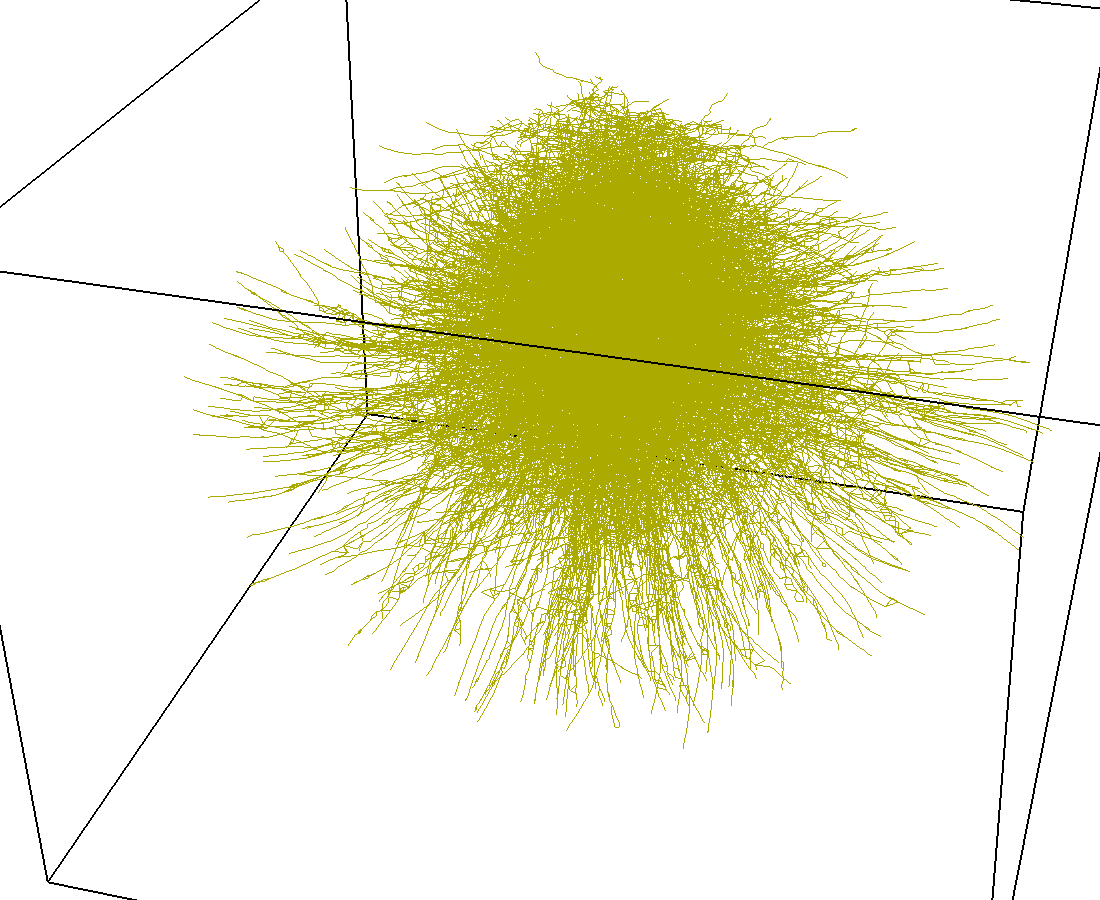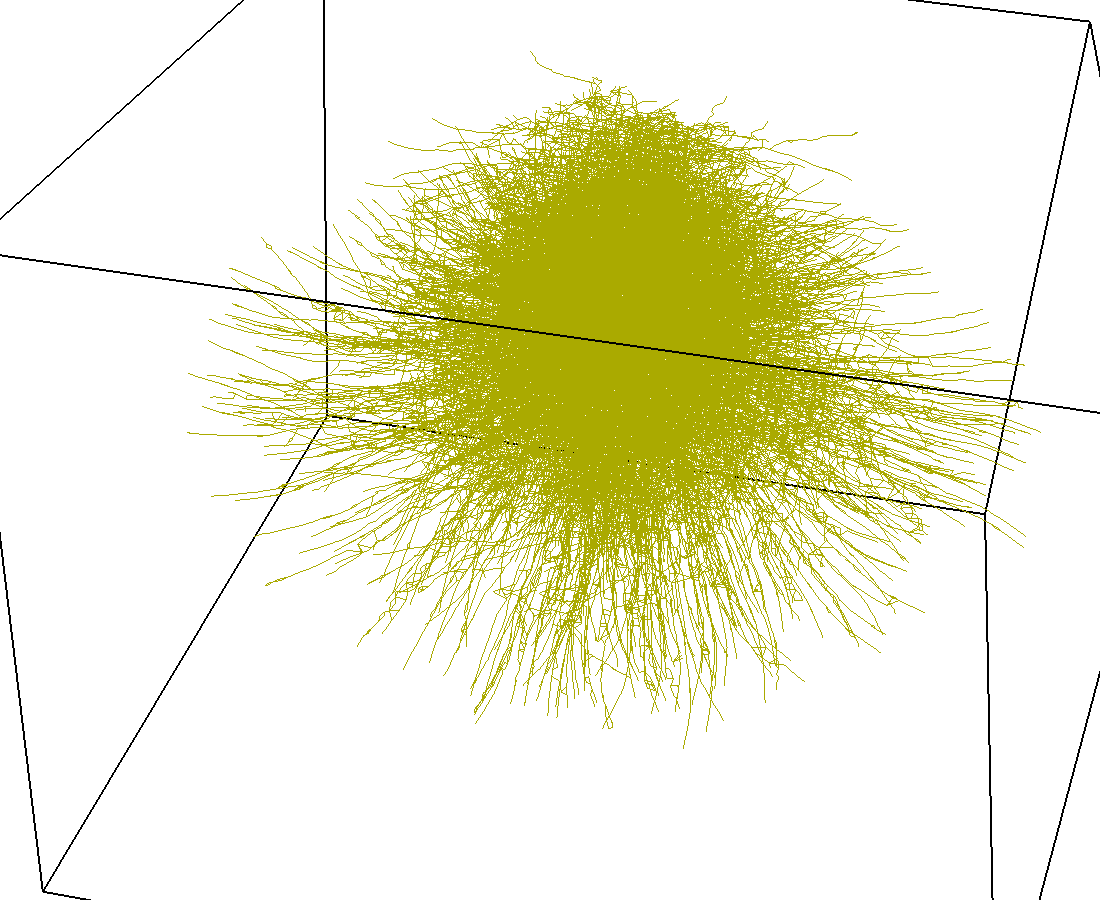
3D model of a brain tissue |
Indexing with 3-level R-tree |
Indexing with 4-level R-tree |
Research Highlights
Improving Spatial Data Processing by Clipping Minimum Bounding Boxes
The majority of spatial processing techniques rely heavily on approximating each group of spatial objects by their minimum bounding box (MBB). As each MBB is compact to store (requiring only two multi-dimensional points) and intersection tests between MBBs are cheap to execute, these approximations
are used predominantly to perform the (initial) filtering step of spatial data processing. However, fitting (groups of) spatial objects into a rough box often results in a very poor approximation of the underlying data. The resulting MBBs contain a lot of “dead space”—fragments of bounded area that contain no actual objects—that can significantly reduce the filtering efficacy. This paper introduces the general concept of a clipped bounding box (CBB) that addresses the principal disadvantage of MBBs, their poor approximation of spatial objects. Essentially, a CBB “clips away” dead space from the corners of an MBB by storing only a few auxiliary points. 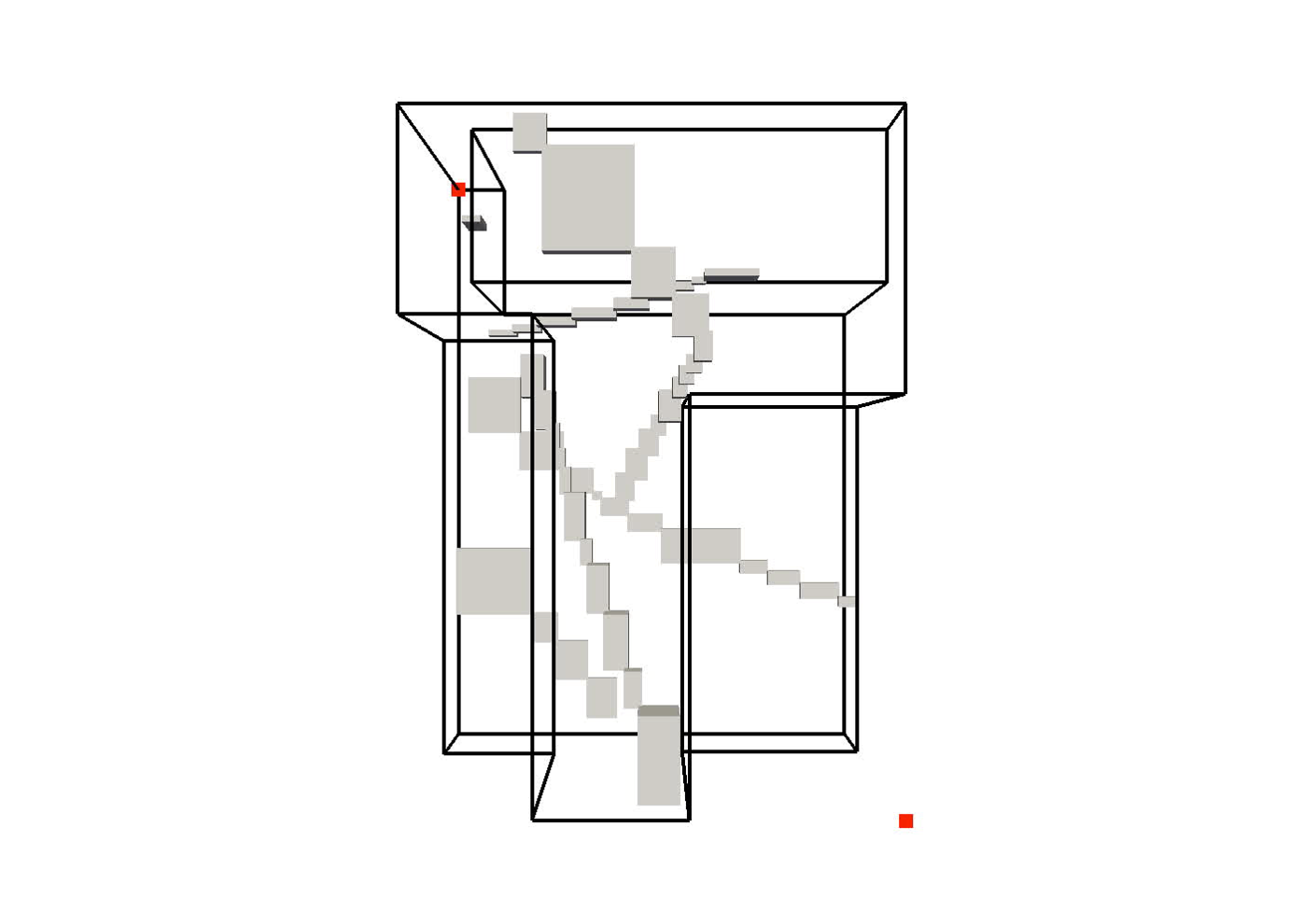
On four popular R-tree implementations (a ubiquitous application of MBBs), we demonstrate how minor modifications to the query algorithm exploit auxiliary CBB points to avoid many unnecessary recursions into dead space. Extensive experiments show that clipped R-tree variants substantially reduce I/Os: e.g., by clipping the state-of-the-art revised R*-tree we can eliminate on average 19% of I/Os. Find out more here…
Efficient Exploration of Scientific Data
Advances in data acquisition—through more powerful supercomputers for simulation or sensors with better resolution—help scientists tremendously to understand natural phenomena. At the same time, however, it leaves them with a plethora of data and the challenge of analysing it. Ingesting all the data in a database or indexing it for an efficient analysis is unlikely to pay off because scientists rarely need to analyse all data. Not knowing a priori what parts of the datasets 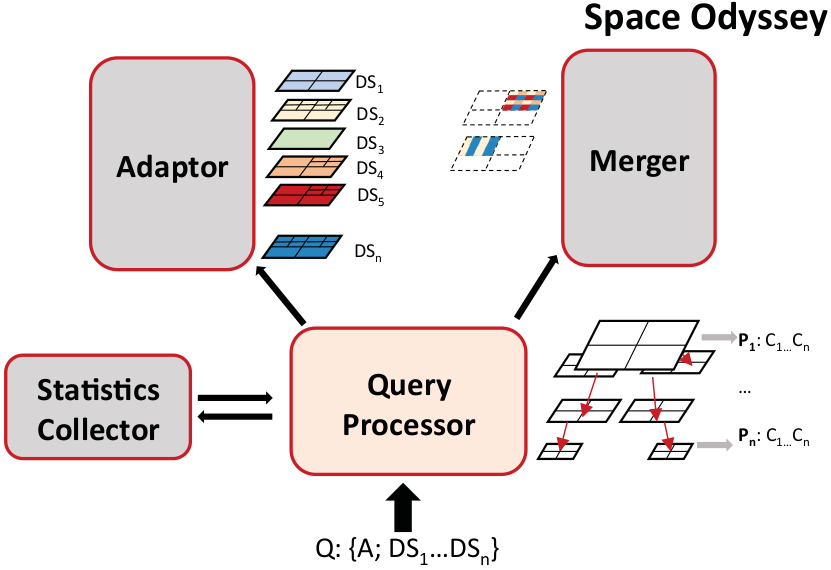 need to be analysed makes the problem challenging.
need to be analysed makes the problem challenging.
Tools and methods to analyse only subsets of this data are rather rare. In this paper we therefore present Space Odyssey, a novel approach enabling scientists to efficiently explore multiple spatial datasets of massive size. Without any prior information, Space Odyssey incrementally indexes the datasets and optimizes the access to datasets frequently queried together. As our experiments show, through incrementally indexing and changing the data layout on disk, Space Odyssey accelerates exploratory analysis of spatial data by substantially reducing query-to-insight time compared to the state of the art. Find out more here…
TRANSFORMERS: Robust Spatial Joins on Non-Uniform Data Distributions:
Spatial joins are becoming increasingly ubiquitous in many applications, particularly in the scientific domain. While several approaches have been proposed for joining spatial datasets, each of them has a strength for a particular type of density ratio among the joined datasets. More generally, no single proposed method can efficiently join two spatial datasets in a robust manner with respect to their data distributions. Some approaches do well for datasets with contrasting densities while others do better with similar densities. None of them does well when the datasets have locally divergent data distributions.
Therefore, we develop TRANSFORMERS, an efficient and robust spatial join approach that is indifferent to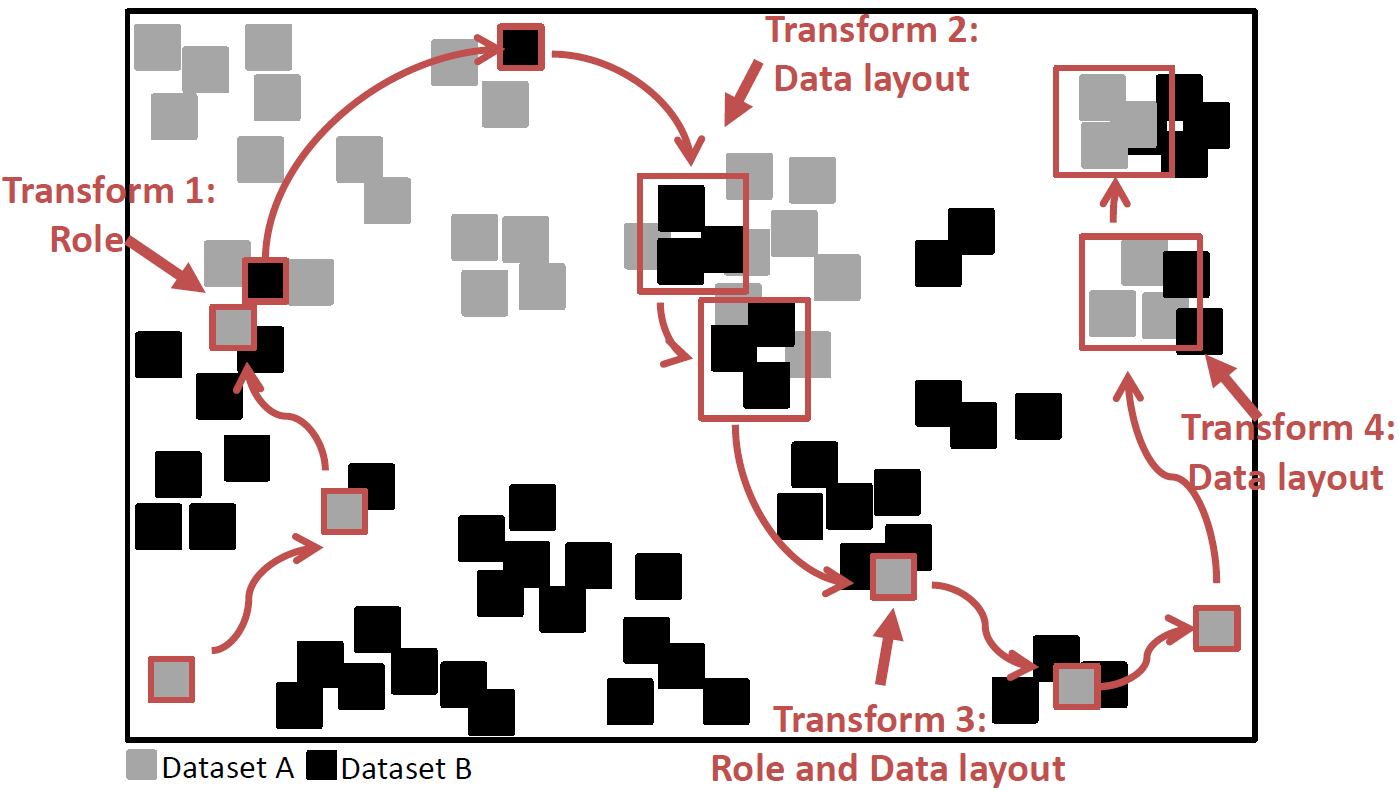 such variations of distribution among the joined data. TRANSFORMERS achieves this feat by departing from the state-of-the-art through adapting the join strategy and data layout to local density variations among the joined data. It employs a join method based on data-oriented partitioning when joining areas of substantially different local densities, whereas it uses big partitions (as in space-oriented partitioning) when the densities are similar, while seamlessly switching among these two strategies at runtime. Continue to read here…
such variations of distribution among the joined data. TRANSFORMERS achieves this feat by departing from the state-of-the-art through adapting the join strategy and data layout to local density variations among the joined data. It employs a join method based on data-oriented partitioning when joining areas of substantially different local densities, whereas it uses big partitions (as in space-oriented partitioning) when the densities are similar, while seamlessly switching among these two strategies at runtime. Continue to read here…
RUBIK: Efficient Threshold Queries on Massive Time Series
RUBIK is a time series indexing tool, which enables scalable threshold queries on a collection of time series. Given the user input, which should be a threshold for the observation value as well as an upper and lower bound for time, and the dataset to be queried, RUBIK returns all the time series values that are above the given threshold for the specified time range.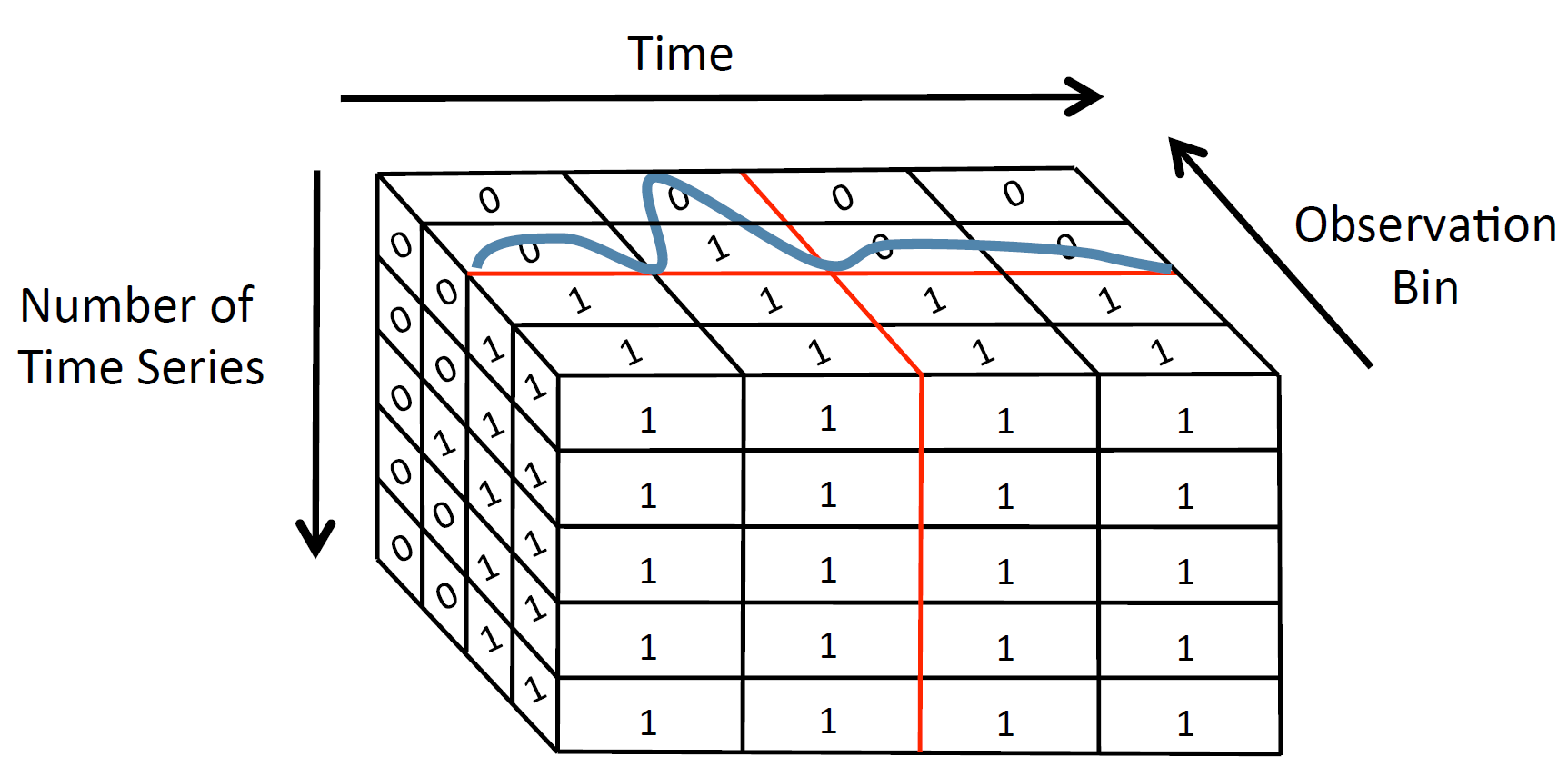
RUBIK transforms threshold queries on time series into a two-dimensional bitmap problem so that queries with time and observation predicates can be efficiently executed as two-dimensional spatial range queries. The size of the bitmap is reduced by applying Quadtree-based decomposition and clustering similar time series.
RUBIK outperforms the state-of-the-art indexing technology for scientific data exploration (FastBit) in answering threshold queries while producing a more space-efficient index. Continue to read here…
FLAT: Accelerating Range Queries For Brain Simulations
Neuroscientists increasingly use computational tools to build and simulate models of the brain. The amounts of data involved in these simulations are immense and the importance of their efficient management is primordial.
One particular problem in analyzing this data is the scalable execution of range queries on spatial models of the brain. Known indexing approaches do not perform well, even on today’s small models containing only few million densely packed spatial elements. The problem of current approaches is that with the increasing level of detail in the models, the overlap in the tree structure also increases, ultimately slowing down query execution. The neuroscientists’ need to work with bigger and more importantly, with increasingly detailed (denser) models, motivates us to develop a new indexing approach.
To this end we have developed FLAT, a scalable indexing approach for dense data sets. We based the development of FLAT on the key observation that current approaches suffer from overlap in case of dense data sets. We hence designed FLAT as an approach with two phases, each independent of density.
Our experimental results confirm that FLAT achieves independence from data set size as well as density and also outperforms R-Tree variants in terms of I/O overhead from a factor of two up to eight. Find out more here…
SCOUT: Prefetching for Latent Structure Following Queries
Today’s scientists are quickly moving from in vitro to in silico experimentation: they no longer analyze natural phenomena in a petri dish, but instead they build models and simulate them. Managing and analyzing the massive amounts of data involved in simulations is a major task. Yet, they lack the tools to efficiently work with data of this size.
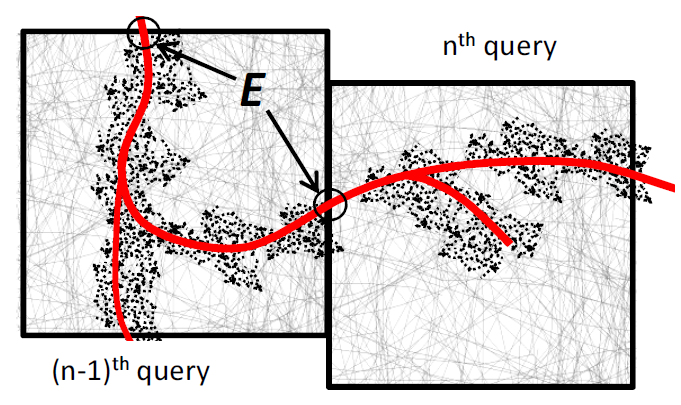
One problem many scientists share is the analysis of the massive spatial models they build. For severaltypes of analysis they need to interactively follow the structures in the spatial model, e.g., the arterial tree, neuron fibers, etc., and issue range queries along the way. Each query takes long to execute, and the total time for executing a sequence of queries significantly delays data analysis. Prefetching the spatial data reduces the response time considerably, but known approaches do not prefetch with high accuracy.
We develop SCOUT, a structure-aware method for prefetching data along interactive spatial query sequences. SCOUT uses an approximate graph model of the structures involved in past queries and attempts to identify what particular structure the user follows. Our experiments with neuroscience data show that SCOUT prefetches with an accuracy from 71% to 92%, which translates to a speedup of 4x-15x. SCOUT also improves the prefetching accuracy on datasets from other scientific domains, such as medicine and biology. Find out more here…
OCTOPUS: Efficient Query Execution on Dynamic Mesh Datasets
Scientists in many disciplines use spatial mesh models to study physical phenomena. Simulating natural phenomena by changing meshes over time helps to understand and predict future behavior of the phenomena. The higher the precision of the mesh models, the more insight do the scientists gain and they thus continuously increase the detail of the meshes and build them as detailed as their instruments and the simulation hardware allow. In the process, the data volume also increases, slowing down the execution of spatial range queries needed to monitor the simulation considerably. Indexing speeds up range query execution, but the overhead to maintain the indexes is considerable because almost the entire mesh changes unpredictably at every simulation step. Using a simple linear scan, on the other hand, requires accessing the entire mesh and the performance deteriorates as the size of the dataset grows.
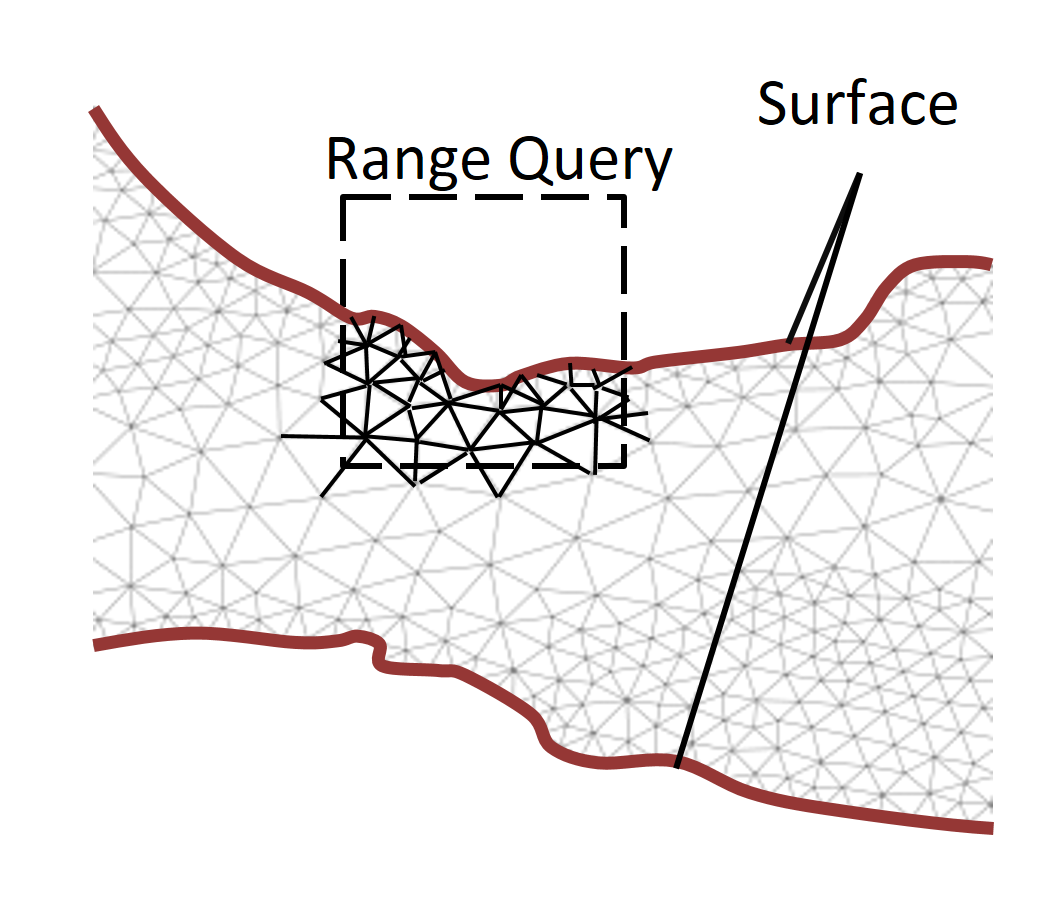
We therefore propose OCTOPUS, a strategy for executing range queries on mesh datasets that change unpredictably during simulations. In OCTOPUS we use the key insight that the mesh surface along with the mesh connectivity is sufficient to retrieve accurate query results efficiently. With this novel query execution strategy, OCTOPUS minimizes index maintenance cost and reduces query execution time considerably. The experiments show that OCTOPUS achieves a speedup between 7.2x and 9.2x compared to the state of the art and that it scales better with increasing mesh dataset size and detail. Continue to read here…
TOUCH: In-Memory Spatial Join by Hierarchical Data-Oriented Partitioning
Efficient spatial joins are pivotal for many applications and particularly important for geographical information systems or for the simulation sciences where scientists work with spatial models. Past research has primarily focused on disk-based spatial joins; efficient in-memory approaches, however, are importantfor two reasons: a) main memory has grown so large that many datasets fit in it and b) the in-memory join is a very time-consuming part of all disk-based spatial joins.
information systems or for the simulation sciences where scientists work with spatial models. Past research has primarily focused on disk-based spatial joins; efficient in-memory approaches, however, are importantfor two reasons: a) main memory has grown so large that many datasets fit in it and b) the in-memory join is a very time-consuming part of all disk-based spatial joins.
To this end we have developed TOUCH, a novel in-memory spatial join algorithm that uses hierarchical data-oriented space partitioning, thereby keeping both its memory footprint and the number of comparisons low. Our results show that TOUCH outperforms known in-memory spatial-join algorithms as well as in-memory implementations of disk-based join approaches. In particular, it has a one order of magnitude advantage over the memory-demanding state of the art in terms of number of comparisons (i.e., pairwise object comparisons), as well as execution time, while it is two orders of magnitude faster when compared to approaches with a similar memory footprint. Furthermore, TOUCH is more scalable than competing approaches as data density grows. Continue to read here…
Demonstrations
Several of our algorithms and indexes have also been presented at conferences as demos. Below, for example, is a YouTube video of the SCOUT demo that highlights and illustrates how SCOUT efficiently prefetches spatial data using the content of previous queries. Videos for FLAT and TOUCH can be found here and here respectively.
Embed of video is only possible from Mediaspace, Vimeo or Youtube
A video showing how the FLAT indexing approach can be used to define and execute queries on the iPad is shown below. Using the well known gestures of iOS, a visualization of the query result can be rotated, zoomed and panned for analysis.
Embed of video is only possible from Mediaspace, Vimeo or Youtube
All the above developed query tools (FLAT, SCOUT, TOUCH, TRANSFORMERS, and RUBIK) are deployed on the PICO supercomputer at CINECA, Italy, as a part of high performance computing tools developed for the Human Brain Project (HBP). The tools are used to efficiently to navigate the simulation datasets produced by HBP. Currently, all the tools are accessed via command line interface and are available only for internal use within HBP. FLAT and SCOUT are also integrated within the BBP SDK.
FLAT usage:
First, the input data set has to be indexed using FLAT:
$ FLATGenerator --inputfile data-file.bin --outputprefix flat
Then, the query file (with spatial queries) can be processed as follows:
$ FLAT --queryfile query-file.txt --outputprefix flat
By default, FLAT outputs the result size of each query to the console while the full result is written to files.
SCOUT usage:
Though SCOUT has a command line interface too (and is more efficient when query file contains spatial query sequences), its main advantages are observed when it is used within the BBP SDK with explaratory user queries (e.g., during visualization). An example of running SCOUT via command line:
$ SCOUT --queryfile query-file.txt --outputprefix flat
Note: user’s data set has to be pre-indexed with FLAT as well.
TOUCH usage:
Given two input files, TOUCH can be used to efficiently find intersecting (touching) points between them. For performance tuning, it supports a number of optional configuration parameters but default values are just fine.
$ TOUCH --datasetA arg --datasetB arg [--logfile filename] [--outfile arg] [--epsilon e] [--runs arg] [--localjoin arg] [--base arg] [--partitions arg] [--partitioningtype arg] --datasetA: filname containing dataset A --datasetB: filename containing dataset B --logfile: log filename (default: SJ.txt) --outfile: output file (default: touches.out) --epsilon: max distance still forming a touch (default: 1.5) --runs: number of runs; for performance measurements (default: 1) --localjoin: local join algorithm used; 0 - nested loop; 1 - plane sweep (default: 1) --base: base (default: 2) --partitions: number of partitions (default: 4) --partitioningtype: type of partitioning; 0 - arbitrary; 1 - hilbert; 2 - x-axis sort; 3 - STR (default: 3)
TRANSFORMERS usage:
While TOUCH is used to efficiently compute intersection points between two spatial datasets in main memory, TRANSFORMERS is an I/O efficient approach, i.e., for datasets that are larger than the available main memory on a computer. The usage is similar:
$ TransformersJoin --datasetA arg --datasetB arg [--resultFile arg] --datasetA: filname containing dataset A --datasetB: filename containing dataset B --resultFile: output file (default: Transformers_Results
Input data for all above algorithms accept the same binary format used by BBP SDK.
RUBIK usage:
First, to import data into RUBIK, use the following command:
$ ImportDataRUBIK --input_data folder_path --output_data folder_path --grid_timesteps nb_timesteps --input_data: folder containing csv files, each file being a cluster of time series --output_data: output binary folder for time series clusters --grid_timesteps: number of timesteps for each time series
Next, you can build RUBIK by running:
$ BuildRUBIK --index_prefix [prefix_path] --source_data [folder_path] \ --grid_timesteps nb_timesteps --grid_voltagebins nb_voltage_bins --index_prefix: output path prefix for the index (each index file will have this prefix) --source_data: binary time series clusters folder (output of ImportRUBIK) --grid_timesteps: number of timesteps for each time series --grid_voltagebins: number of voltage bins used (must be power of two)
Finally, times series can be queried as follows:
$ QueryRUBIK --index_prefix prefix_path --start_time start_time --end_time end_time \ --voltage-gt voltage_value --grid_timesteps nb_timesteps \ --grid_voltagebins nb_voltage_bins --multiple_runs nb_runs -i,--index_prefix: path prefix for the index (output of BuildRUBIK) -t,--start_time: lower time bound for query -u,--end_time: upper time bound for query --voltage-gt: lower voltage bound for query --grid_timesteps: number of timesteps for each time series --grid_voltagebins: number of voltage bins used (the same as in building) -s,--multiple_runs: define how many times the query should be executed (for performance measurements only)
External Funding Sources
This work is supported by the Hasler Foundation (Smart World Project No. 11031) and the European Union FET Flagship “Human Brain Project”.
People involved: Eleni Tzirita Zacharatou, Mirjana Pavlovic, Thomas Heinis, Darius Šidlauskas, Lionel Sambuc
Publications
Please note that the publication lists from Infoscience integrated into the EPFL website, lab or people pages are frozen following the launch of the new version of platform. The owners of these pages are invited to recreate their publication list from Infoscience. For any assistance, please consult the Infoscience help or contact support.