Decoding the Immunological Microtubule Network
|
Improving vaccination and cancer immunotherapy are key areas of active biomedical research. Both processes are fundamentally linked by the shared requirement for optimal leukocyte activation and motility. Despite its fundamental importance, the study of leukocyte infiltration and interactive behavior has received limited attention from immunologists and clinicians. Importantly, the role of the microtubule (MT) network in these processes has so far largely been ignored. |
 |
|
In fact, the MT network is important for a host of key cell functions. Importantly, the role of the microtubule (MT) network in these processes has so far largely been ignored. In fact, the MT network is important for a host of key cell functions. The dynamic and mechanical properties of MTs are regulated via the incorporation of tubulin isotypes and post-translational modifications (PTMs), forming a ‘tubulin code’. The resulting structural and chemical variety is essential for the control of the dynamics and mechanical stability of the tubulin network. Importantly, dysregulation of mechanisms involved in the tubulin code can result in disease. |
Project goals: |
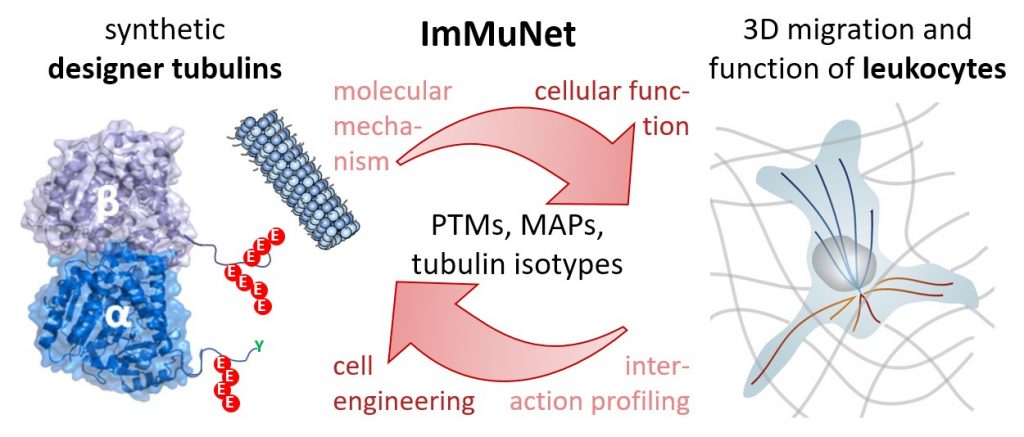
|
In this project, we explore the MT regulatory axis from a molecular to tissue level perspective using approaches that span the entire range from tubulin chemistry to analysis of organismal immunology. We investigate: 1. How do PTMs alter tubulin stability and dynamics?2. Which microtubule associated proteins (MAPs) read the modified MT network in immune cells?3. How is immune cell tissue invasion and function controlled by the tubulin code? |
Open positions
We are looking for motivated PhD students and postdocs.
If you are interested, please contact the committee members directly.
Protein chemistry and Chemical Biology: beat.fierz@epfl.ch
Biochemistry and Cell biology of microtubules: charlotte.aumeier@unige.ch
Cellular Morphodynamics: michael.sixt@ist.ac.at
Adaptive Immunity: jens.stein@unifr.ch
Project publications:
Tubulin engineering by semisynthesis reveals that polyglutamylation directs detyrosination, Ebberink, E., Fernandes S., Hatzopoulos G., Agashe N., Guidotti N., Reichart T.M., Reymond L., Velluz M.-C., Schneider F., Pourroy C., Janke C., Gönczy P., Fierz B., Aumeier C., 2022 bioRxiv; doi: https://doi.org/10.1101/2022.09.20.508649



