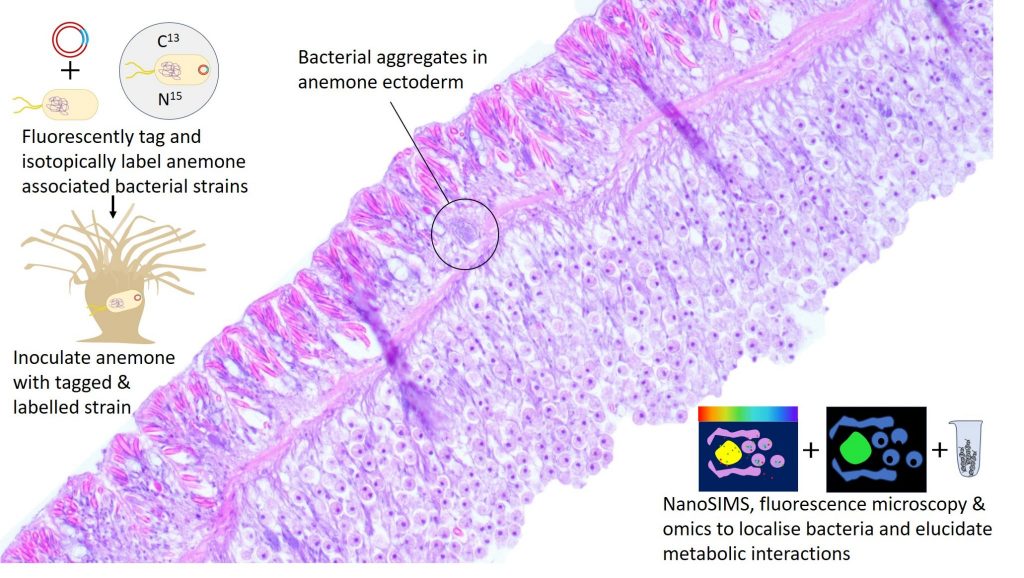
Elucidating bacterial metabolic contributions to the cnidarian holobiont using a genetic transformation approach
Cnidarian holobionts comprise the cnidarian animal and it’s associated symbiotic microbes, including algae, archaea, fungi, viruses and the focus of this project: bacteria. These organisms contribute to holobiont functioning and fitness, for example by participating in nutrient acquisition and metabolic exchange.
This project aims to determine metabolic roles of specific bacterial taxa and the fine scale localisation of these symbionts within the holobiont. We use bacterial strains isolated from wild Mediterranean Aiptasia mutabilis, a model photosynthetic cnidarian, to further our understanding of the mechanistic interactions of cnidarian-associated bacteria with both the host and the symbiotic algae. We combine experimental microbiome manipulation, genetic transformation, correlative imaging and NanoSIMS, and omics in a multipronged approach.
