Development of computational design methodologies

Due to limited understanding of the principles that govern protein function and structure, the design of functional proteins remains challenging. New computational design methodologies are a powerful tool to engineer novel proteins. Our laboratory develops and applies novel computational methodologies to create functional proteins with therapeutic potential. The computationally generated protein sequences are then tested by the experimental arm of the laboratory, providing an integrated pipeline of theoretical and experimental work.
Bottom-up de novo protein design
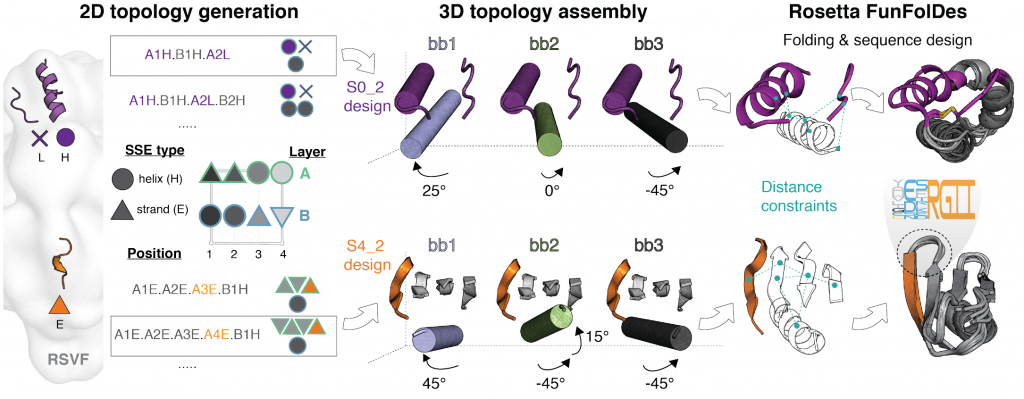
We have developed TopoBuilder, a new “bottom-up” strategy for de novo protein design. In contrast to most protein design methods, the local structural and global topological requirements are optimized to present and stabilize desired functional motifs. The TopoBuilder algorithm executes three steps (see Figure 1):
- 2D topology generation – Enumeration of protein topologies compatible with the functional motif in 2D space.
- 3D topology assembly – Projection in 3D space by assembling idealized secondary structure elements (SSEs) around the fixed functional motif. These 3D structures, referred to as “sketches,” are further refined using parametric sampling.
- Iterative folding and sequence design – Refinement of structural features coupled with sequence design using Rosetta FunFolDes.
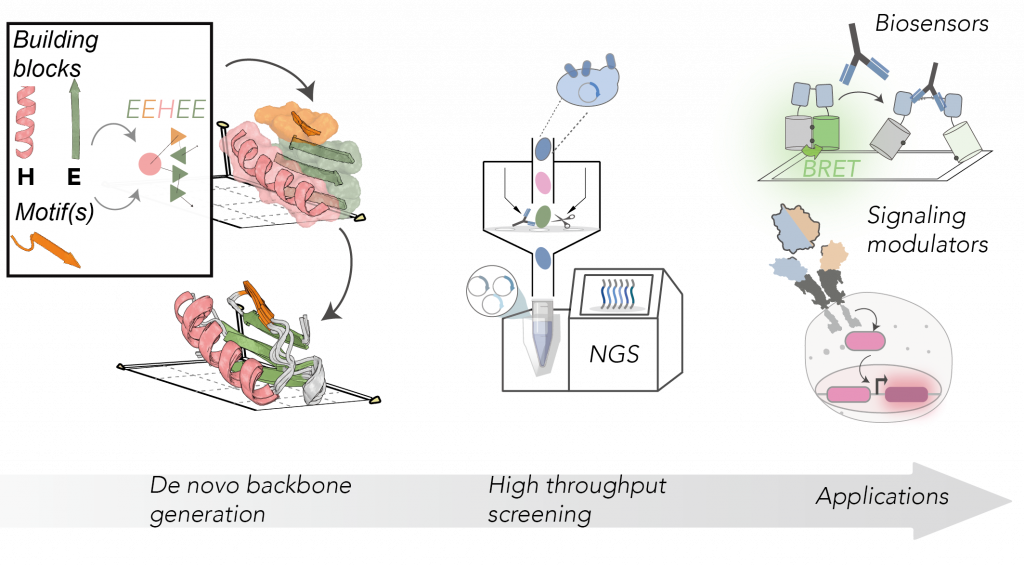
The obtained protein sequences are encoded into libraries and displayed on the surface of yeast cells. Using a Next Generation Sequencing (NGS) readout, we quantify the functional features of each sequence in a high-throughput fashion. The designs can perform complex biochemical functions, such as monitoring antibody responses and modulating synthetic signaling receptors.
References: Yang C. et al. Nature Chemical Biology (2021), Sesterhenn F. et al. Science (2020), Bonet J. et al. PLoS computational biology (2018)