Development of drugs based on cyclic peptides
Most drugs function by binding to and modulating protein targets. For many targets however, it has been challenging to develop drugs based on “classical” small molecules (orange in figure below) because the proteins do not contain pockets or clefts for binding. Macrocyclic molecules such as cyclic peptides (green) or small cyclic peptides/macrocyclic compounds (blue) can bind to relatively flat, featureless surfaces, and thus to challenging targets. The good binding properties of macrocyclic molecules stems to a large extent from the cyclic configuration and the resulting limited flexibility, which reduces the entropic penalty upon target engagement. In our laboratory, we are developing and applying methods for generating and screening large and structurally diverse libraries of larger and smaller cyclic peptides, and we aim at developing the identified ligands into therapeutics.
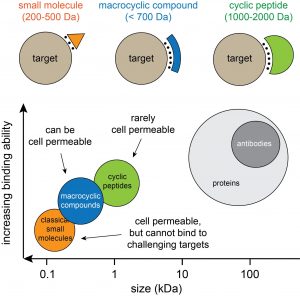
Figure: Comparison of different drug modalities. Cyclic peptides (1-2 kDa) can be developed efficiently by biological display methods such as phage display. These techniques allow the facile generation and screening of billions of different cyclic peptides. The larger cyclic peptides typically do not cross membranes easily and are thus developed against extracellular proteins. Smaller macrocyclic molecules such as small cyclic peptides (< 1 kDa) are harder to develop. However, if they have a small polar surface, they can passively diffuse into cells and can be used to develop drugs against intracellular proteins.
Publications:
Carle, V., et al., Journal of Medicinal Chemistry (2021)
Gonschorek, P., et al., Journal of Medicinal Chemistry (2022)
Phage display selection of bicyclic peptides
We routinely generate bicyclic peptide ligands to disease targets using a phage display-based strategy shown in the figure below. In brief, cysteine-rich peptides displayed on phage are reacted with a small organic linker to form bicyclic peptide structures. Large combinatorial libraries (> a billion variants) of bicyclic peptides (bound to phage) are isolated in affinity selections. Binders are identified by sequencing the encoding DNA that is enclosed in the phage particle.
A long-standing goal of the pharmaceutical industry has been the development of peptides that can be taken as a pill. Usually, peptides are immediately degraded by proteases in the stomach and intestines. We have recently developed a cyclic peptide format (double-bridged peptides) and a method that allowed the in vitro evolution of highly stable peptides. We have shown that these peptides are not degraded by proteases upon oral administration.
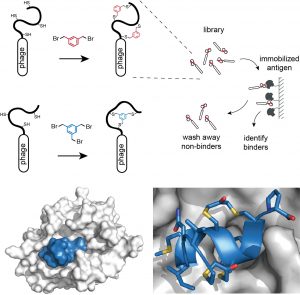
Figure: Strategies for chemically cyclizing phage-encoded peptide libraries and X-ray structure of a double-bridged peptide developed by phage display.
Publications:
Heinis, C., et al., Nature Chemical Biology, 5 (2009)
Zorzi, et al., Nature Communications, 8 (2017)
Kale, S., et al., Nature Chemistry, 10 (2018)
Wilbs, J., et al., Nature Communications, 11 (2020)
Kong, X.-D., et al., Nature Biomedical Engineering, 4 (2020)
Diaz-Perlas, C., et al., Nature Communications, 14 (2023)
High-throughput screening of macrocycles
We are interested in generating macrocyclic drugs that can be applied orally and/or that are cell permeable to target intracellular proteins. Toward this end, we are developing methods to generate and screen large combinatorial libraries of small cyclic peptides (< 1 kDa). Our strategy is to synthesized the cyclic peptides by combinatorially dispensing building blocks to wells of microtiter plates so that they can react and form macrocyclic structures. We are using efficient reactions to obtain macrocycles at good yields and reasonable purity, to enable screening in the same microwell plates. The omission of purification steps allows us to generate and screen large libraries of ten-thousands of molecules.
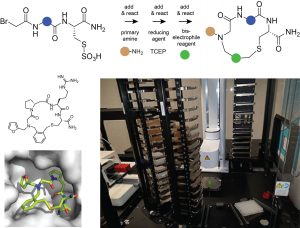
Figure: Strategy used for synthesizing small cyclic peptides and robotic platform used for the synthesis and screening.
Publications:
Kale, S., et al., Science Advances, 5 (2019)
Mothukuri, G.K., et al., Chemical Science, 11 (2020)
Sangouard, G., et al., Angewandte Chemie Int. Ed. (2021)
Habeshian, S., et al., ACS Chemical Biology (2022)
Habeshian, S., et al., Nature Communications (2022)
Bognar, Z., et al., Organic & Biomolecular Chemistry (2022)
Merz, M.L., et al, Nature Chemical Biology (2023)
Schüttel, M., et al, Journal of Peptide Science (2024)