Research
Our Mission
Obesity and its related morbidities are multifactorial disorders with a heavy cost on population health. Obesity is characterized by an abnormal fat accumulation resulting from whole-body energy imbalances. By investigating the molecular basis by which metabolites, including bile acids and other signaling molecules, act as controllers of energy homeostasis, our laboratory aims to identify novel mechanisms and strategies to target a wide range of chronic disorders, including metabolic disease and cancer.Research areas and approaches
The liver–gut–brain axis is a physiological system specialized in fuel sensing and processing. Messenger molecules of diverse chemical composition and origin mediate cross-talks between these three organs. Our research aims to focus on this system and gain insight into the mechanisms by which nutrient-derived metabolites in general and bile acids, in particular, are sensed and integrated with the hormonal and neuronal system to coordinate overall energy utilization and homeostasis. At the subcellular level, our research indicates that mitochondria play a key role in mediating these responses. Understanding how these organelles are regulated is thus at the center of our research.
The liver – gut axis
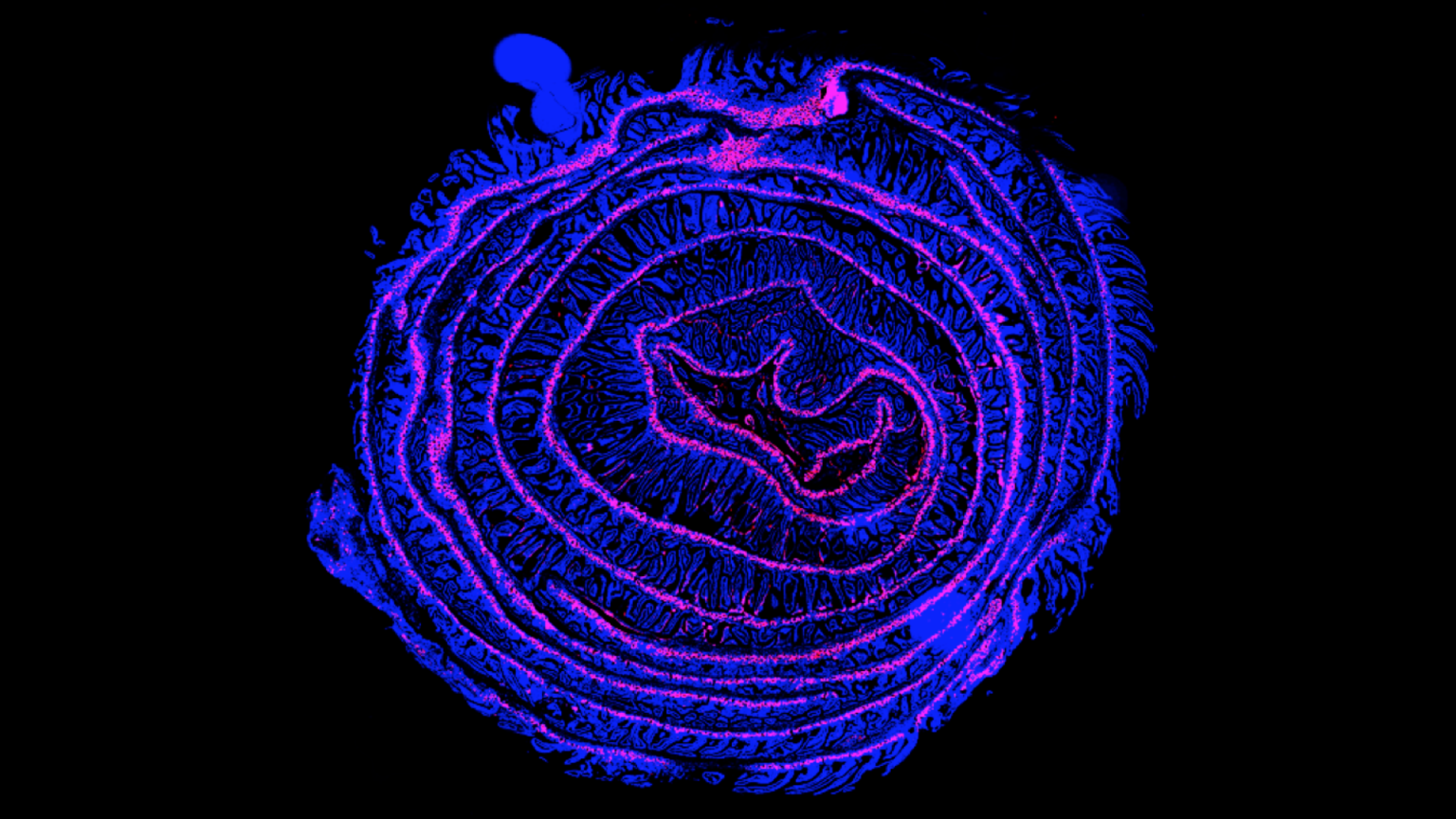
The liver-gut axis is a system specialized in nutrient sensing and processing that plays a major role in the regulation of energy intake. These two organs communicate extensively via the biliary tract, portal vein, and systemic mediators. Intestinal factors regulate bile acid synthesis, glucose and lipid metabolism in the liver whereas hepatic molecules, such as bile acids, influence intestinal homeostasis. Our research covers both ways of this communication system. By investigating how nutrient-derived molecules activate hepatic nuclear receptors, we study the regulation of hepatic metabolism by the intestine and the gut microbiome. Conversely, we are interested in understanding how liver-produced bile acids can control intestinal renewal under normal and pathological conditions. This research is performed using state of the art in vivoapproaches involving genetically modified animals as well as ex vivointestine and liver stem cell-based organoid culture system. Such organoids self-organize into 3D matrigel environment and recapitulate tissue architecture and hepatic as well as intestinal metabolic functions. These organoids are well-suited for genetic manipulation, imaging, drug screening as well as genomic/proteomic/metabolomic analysis and constitute excellent tools to investigate fundamental questions related to the liver-gut axis and its regulation by signaling molecules. Our team also uses intestine and liver organoids to model metabolic dysfunctions, such as non-alcoholic fatty liver disease and intestinal cancers.
The brain – liver axis
The brain-liver axis has traditionally been studied in the context of stress induced by a lack of nutrients. During prolonged fasting, the brain coordinates responses between the liver and different fat depots to ensure that fuel is provided in the form of sugar and ketone bodies to vital organs. Our lab pushes these boundaries by investigating the role of this axis in the context of controlled energy expenditure triggered by other types of stress such as hypothermia. At the anatomical level, we study how projections from the sympathetic nervous system innervate the liver and regulate hepatic metabolic pathways linked to the control of whole-body energy homeostasis. At the cellular level, we investigate the respective contributions of pericentral and periportal hepatocytes to liver homeostasis and energy output. Finally, at the sub-cellular levels, our work is focused on hepatic mitochondria and how a set of liver-specific transporters regulate hepatic fuel utilization in the context of energy expenditure. We also study how hepatic metabolites, such as bile acids, impact on central regulation. In particular, how these molecules inform the hypothalamus about the nutritional state of the organism. To unravel these novel functions, we are using state-of-the-art approaches in biochemistry, molecular biology, and mouse genetics. An integrative approach combining genetically engineered mouse models, metabolic phenotyping and in-depth cellular and molecular profiling in cellular and ex vivo organoid models is used to reconstruct the networks that are modified by hormonal and neuronal signaling molecules.
System genetics of bile acid signaling
Given the powerful beneficial actions of bile acids as hormones acting on multifactorial metabolic diseases and aging, our lab is interested in understanding the genetic and environmental factors that control BA homeostasis. To this end, we have set up an unbiased systems genetics approach to identify genetic and environmental determinants that control BA signaling and to define the impact of BAs on health and disease. This approach relies on the analysis of a diverse panel of recombinant inbred mouse lines that descend from crosses between a C57BL/6J mother and a DBA/2J father—also referred to as BXD mouse genetic reference population (GRP). Our lab has used this GRP to collect normative and experimental clinical phenotypes and developed strong bioinformatics skills to correlate changes in BA levels with metabolic parameters and phenotypes. Computational and bioinformatics approaches are being applied not only to our system genetics approach but to all the above-mentioned projects for which we are using omics methodologies (metabolomics, proteomics, transcriptomics, and epigenomics).