In SuNMIL we combine fundamental science research on solid-liquid interfaces with inspiration from Nature to create new molecules and new materials for a variety of applications. SuNMIL’s approach to research is guided by two principles that together constitute its mission.
We try to develop novel fundamental science in the field of solid liquid interfaces. In particular, we aim at developing techniques so to advance the knowledge on the fundamental problem of proteins in water.
At the same time, we have the ambition to tackle great unresolved challenges for mankind. Currently we are focusing on two problems. The first is the development of broad-spectrum antivirals. The second is an attempt to generate more sustainable polymeric materials.
At SuNMIL, we like to focus on problems that plague under-developed or developing countries. For example, millions of people die every year because of viral diseases, most in developing countries. For this reason, we developed a line of research on supramolecular approaches to design novel antivirals. For the same reason, our approach to generating recyclable plastics is rooted on the idea of generating value from waste.
As explained above, in our laboratory, the research themes are broad, similarly all forms of diversity are welcomed and fostered we are simply linked by an honest enthusiasm for science in all of its forms.
Biological Application
Environmental Application
Nanoparticle Studies

Protein Research
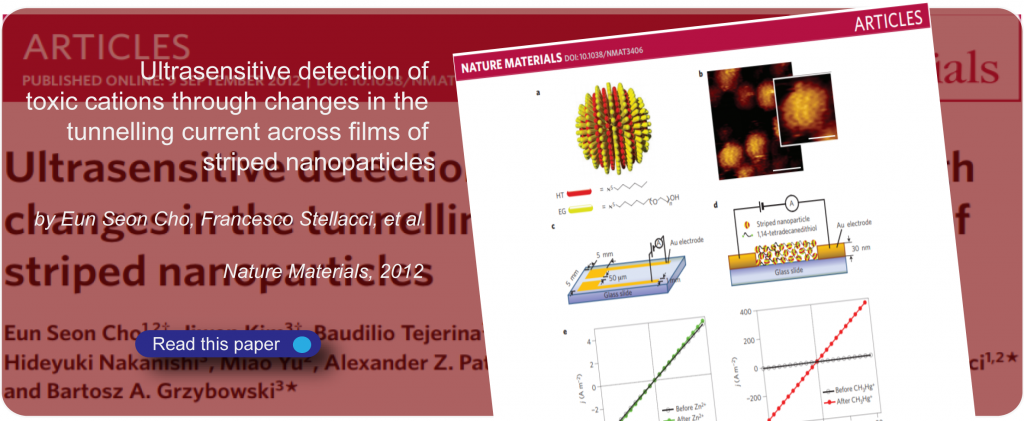
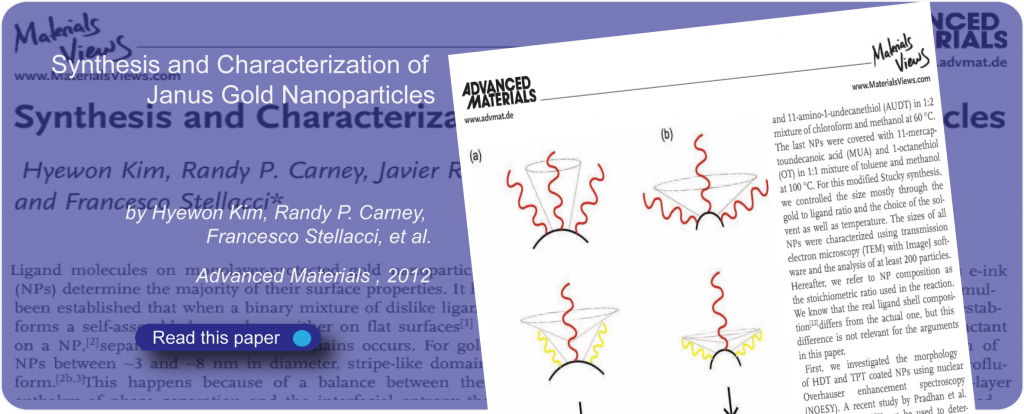
Proteins form the basis of life, and interactions between proteins and molecules are linked to the organization and communication of cells, as well as to effects that influence our daily lives.
Weak interactions are responsible for many complex protein functions.
At SuNMIL we aim to fundamentally understand how protein interactions can be modulated in the presence of small molecules. We use approaches that we borrow from material science, physical chemistry, and biology to unveil the mechanism at the basis of this interaction as well as to discover novel biological functions for small molecules.
Here you can watch a descriptive video prepared by our group.
Ting Mao, Ph.D. student
Understanding protein-protein interaction and how certain small molecules regulate it is of great fundamental importance in physiology and pathology. However, due to the complexity of protein surface topologies, direct measurement of interactions on proteins varies across protein systems, which adds to the difficulty of the problem. In this project, I demonstrated a stable nanoparticle system where I can control the surface components to incorporate interactions that exist in proteins to study the colloidal part of the protein interaction problem.
Ekaterina Poliukhina, Ph.D. student
My work focuses on two interconnected aspects: developing an experimental approach utilizing cryogenic electron tomography to extract inter-protein interaction potential (IP) and modeling the obtained under different systems conditions IP to retrieve its universal decomposition form.
Cécilia Laetitia Carla Siri, Ph.D. student
I am interested in 1) investigating the effect of proteinogenic amino acids on protein-ligand interactions and 2) deciphering the role of peptides as modulators of protein-protein interactions. In my work, I use different analytical techniques from chemical biology, such as ITC, NMR, and HPLC-MS.
Dr. Quy Ong Khac, Senior Scientist
I am the senior scientist in the group and I follow all projects. Recently, I have invented a method that from cryo-electron microscopy images leads to the potential of mean force of the objects images. This method can be used for nanoparticles and also for proteins.
Dr. Pamina Winkler, Post-Doctoral Fellow
My overarching research goal is to quantify the influence of proteinogenic amino acids on protein cross-interactions. To achieve this, I apply a versatile tool set from physical chemistry and biology to obtain quantitative values such as the binding affinity of protein cross-interactions. I am interested in gaining a deeper insight into the underlying mechanism at play by further combining my experimental approach with AI-based computational protein design.
Dr. Xufeng Xu, Post-Doctoral Fellow
By using analytical ultracentrifugation and performing cell experiments, I want to understand the roles of proteinogenic amino acids in 1) changing protein-protein interaction (fundamental physical chemistry) and 2) modulating protein phase separation in cells (molecular cell biology).
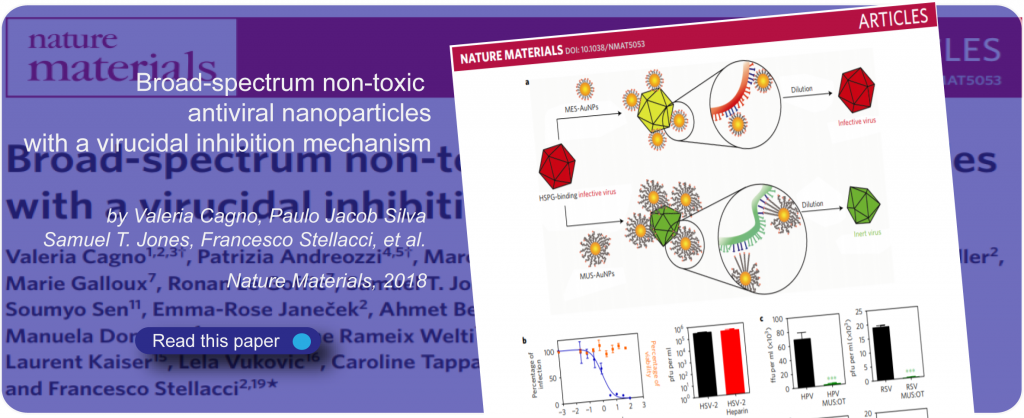
Antivirals
Viruses cause untold damage to human health and well-being. Many viruses that cause human disease are without effective antiviral drugs to treat the infections they cause, meaning that treatment options are often limited during outbreaks. The limited arsenal of currently-approved antiviral drugs is primarily deployed to target and interfere with the ability of viruses to replicate their genome within host cells. These drugs often impart off-target effects, toxicity and the emergence of drug resistance in virus populations.
In pursuit of broad-spectrum antiviral activity, SuNMIL’s current scope of work includes both enveloped and non-enveloped viruses, with representative members from both the DNA and RNA viruses world. In spite of the enormous genotypic and phenotypic diversity among human viruses, commonalities exist in how they engage with the target cell prior to establishing infection, converging on a relatively small number of macromolecules that facilitate viral attachment to the cell and preclude virus entry.
At SuNMIL, we seek to hijack the attachment of viruses to host cells by mimicking these macromolecules found at the cell surface. When successful, we create molecules that are efficient at binding to viruses and can block their entry. While this approach has been known for years, it has not lead to successful therapeutic agent because after associating to viruses, entry inhibitor loose efficacy upon dilution of drug. We have discovered that this limitation can be overcome by designing the molecules so that, after binding, they are able to destroy the viruses before they can engage with the target cell. The SuNMIL virology team combines expertise in microbiology, molecular biology, chemistry and materials science to develop antiviral compounds against a wide range of human pathogens and improve our ability to combat emerging and existing viral threats.
Yong Zhu, Ph.D. student
I am targeting Heparan Sulfate Glycans (HSPG), the virus’s initial binding site on host cells, in quest to create molecules resembling HSPGs. The goal is to irreversibly neutralize viral particles before they infect host cells. My research identified key structural features for potent antiviral effects, with a lead molecule exhibiting outstanding efficacy against various viruses, including SARS-CoV-2, without causing harm to cells. This molecule also reduced SARS-CoV-2 infection in hamsters similarly to the FDA-approved antiviral nirmatrelvir. My next steps involve a comprehensive exploration of the antiviral mechanism.
Francesca Olgiati, Ph.D. student
The world has changed during the last years, due to pandemic caused by the spread of SARS-CoV-2. We are always fighting against viruses, some that are new and emerging, but also some others that have existed for hundreds of years and are still causing medical and economic impacts. For these reasons, it is important to develop new antiviral drugs that are broad-spectrum and can be adapted to many different viruses with minimal modifications.
Our group develops macromolecules that can establish interactions with viruses and inhibit their infectivity through a permanent disruption of the viral membrane.
These macromolecules are based on gold nanoparticles or b-cyclodextrins and we chemically modify them according to the virus and type of interaction we want to establish.
Specifically, in this project, we work with protein-protein interactions and use peptides to bind viruses. In particular, we focus on influenza viruses and peptide-based beta-cyclodextrins that lead to irreversible in-vitro (on cells) inhibition of the virus.
Anita Zucchi, Guest Ph.D. student from Università degli Studi di Milano-Bicocca, Italy
The aim of this project is the synthesis of original and innovative molecular broad-spectrum antivirals and their characterization. The synthetic targets are characterized by the presence of iodinated aromatic core -such as Iopamidol whose biocompatibility is already known given its well-established use as X-ray contrast agent in diagnostics. Ensuring minimal to no toxicity is crucial for achieving optimal biocompatibility and preventing undesirable side effects. Therefore, leveraging an FDA-approved, essentially non-toxic compound provides a significant advantage.
Iopamidol and the related compounds are scaled up materials, widely available on the market thus constituting an economic raw material for the preparation of the construct. Moreover, the presence of the iodine could be exploited to carry out imaging statues using the antiviral construct as a contrast agent as well.
Dr. Frank William Charlton, Post-Doctoral Fellow
In June 2023 I brought my virology background to SuNMIL to define the mechanism(s) of action of our current repertoire of antiviral compounds. Aside from this, my work also involves the expansion of our compounds’ antiviral activity to additional viruses of clinical significance. As part of a multidisciplinary team, it is my role to offer expertise in virology whilst working alongside scientists of varying backgrounds to characterize and deliver novel antiviral compounds.
Dr. Sujeet Pawar, Post-Doctoral Fellow
I am a synthetic organic chemist whose research focuses on developing broad-spectrum antivirals. To do that, I design and create new carbohydrate-based inhibitors against viral proteins to further test them to study efficacy and safety in preclinical settings. I also work in biomaterials to address therapeutic gaps in nerve regeneration by introducing novel protein activators which are carbohydrate-based small molecules in the polymeric scaffold.
Dr. Rémi Andréa La Polla, Post-Doctoral Fellow
My project is to study in depth the interaction of our antiviral molecules with host cells and to develop molecules that can facilitate viral entry.
Dr. Paulo Henrique Jacob Silva, Post-Doctoral Fellow
My research focuses on translating anti-influenza molecules from the laboratory to the clinics. I follow the whole process from optimizing the synthesis to designing in-vivo experiments.
Dr. Shadi Kordbacheh, Post-Doctoral Fellow
The goal of my project is introducing novel antiviral drugs to target viruses virucidally. My focus is on the synthetic part of the drug introduction and finding new peptides which can destroy viruses permanently. These peptides can be conjugated to variety of cores and hydrophobic chains or can be utilized individually as long as they have virucidal effect.
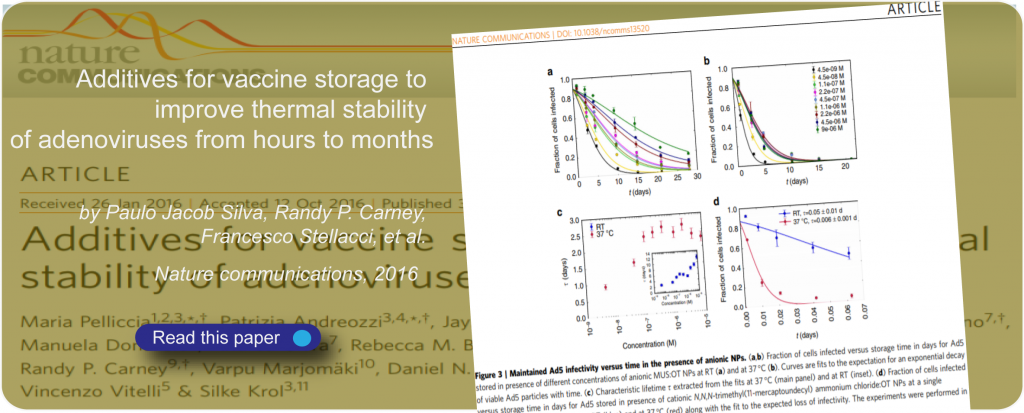
Nature-inspired circular recycling (NaCRe)
Nature-inspired circular recycling (NaCRe) is an approach developed in SuNMIL to address the challenge of recycling polymeric materials.
As the name states, it originates from the way nature recycles its polymers, in particular, proteins. In nature, multiple types of proteins can be degraded into their monomers, the amino acids, which then can be reused to build new proteins that have little to nothing in common with the initial proteins. The key to this ideal recycling approach, that starting from a random mixture can lead to any protein known on earth is the nature of proteins themselves. They are sequence-defined polymers whose structure (and consequently function) depends on the exact sequence of the twenty proteogenic amino acids that compose them. A newly synthesized protein is “just” a scrambled version of many proteins that existed before.
Our goal is to understand the potential of this recycling approach and to use to either strengthen the case for proteins to be used as ideal sustainable materials or to learn from them and generate truly sustainable soft polymeric materials.
Our team is dedicated not only to protein recycling but also to the production of materials derived from proteins. The aim is to create materials with properties similar to those of plastic.
We are a multidisciplinary team, which explores NaCRe from the biological, materials science and chemistry point of view.
Here you can watch a descriptive video prepared by our group.
Laura Roset Julia, Ph.D. student
We have recently shown in our lab that soluble proteins can be recycled into new proteins in vitro. The goal is now to reach the milligram scale for materials, meaning the synthesis of a protein material made out of amino acids form a previously degraded protein material different from the final one. This process involves materials’ solubilization, protein digestion, recombinant protein expression, and protein materials’ engineering. With this, we aim to prove that NaCRe can be brought to the milligram scale and provide a circular recycling scheme for protein materials, while showing that sequence-defined polymers could be a strategy to achieve sustainability of soft materials.
Anna Koptelova, Ph.D. student
I am a Ph.D. student at SuNMIL and Sustainable Materials Laboratory .
My research focuses on layering various waste proteins to improve packaging properties. Our goal is to determine how we can create a recyclable, bio-based multicomponent or multilayer material with qualities comparable to fossil-based polymers. The primary properties under investigation include water and oxygen barriers, as well as mechanical strength. Ongoing efforts and future directions involve studying the characteristics of single-layer protein materials and then identifying the optimal method for creating a multilayer material with enhanced properties.
Chihui Zheng, Ph.D. student
My research focuses on establishing a recycling system for polymeric materials formed by commercially available monomers. By exploring the conditions for selective degradation, we are reaching the selective recovery and separation of various polymers. This aims to create sustainable soft polymeric materials with different functionalities and applications by combining polymers with different properties or forming copolymers from different monomers.
Dr. Ivana Ivancova, Post-Doctoral Fellow
Recycling of water insoluble proteins brings new challenges to the concept of protein waste recycling. Meat industry produces huge amounts of keratin waste (which is 95-98% protein bioresource). The strong structure of the keratin, indigestibility by most proteases and insolubility in water, weak acids and alkali are the main obstacles on the way to its recycling. In our lab, we try to develop an effective recycling method of the wool to establish this protein rich waste as a sustainable protein/peptide/amino acid source.
Dr. Youwei Ma, Post-Doctoral Fellow
My research is centered on advancing the design of high-performance polymers with recyclability, particularly by utilizing food protein waste as a primary feedstock. The objective is to enhance the strength and toughness of protein-based polymers, with the ultimate goal of manufacturing a fully biodegradable protein polymer—whether in the form of plastic or elastomer—as a sustainable replacement for its synthetic, non-recyclable Counterparts.
Dr. Gadi Slor, Post-Doctoral Fellow
While it is clear that NaCRe principles can be applied for biopolymers, we believe that parts of it can be applied to synthetic polymers (plastics) as well. One of the biggest challenges of plastic recycling is the inability to recycle waste streams that contain mixtures of different plastics, forcing a process of sorting and separating prior to recycling. In many cases, among them in multilayered packaging, sorting is not possible and therefore they are being landfilled or incinerated. My research is focusing on the discovery and characterization of new recyclable synthetic polymers, and using them to create a new paradigm on plastic recycling that enables closed-loop recycling of complex plastic mixtures.
Other Research Interests at SuNMIL
Emie-Kim Ngo Tan, Ph.D. student
mRNA delivered with lipid nanoparticles (LNPs) hold huge potential for next-generation therapeutics. For the targeted in vivo delivery of LNPs, we need to upgrade our understanding on the relationship between LNPs’ key structures (ie, composition, size, surface ligands, structural dynamics etc.) and their biodistribution.
Heyun Wang, Ph.D. student
My project started with developing antiviral surfaces and now it has morphed into studying approaches to efficiently produce lipid nanoparticles.
Melis Özkan, Ph.D. student
I am a Ph.D. student at SuNMIL and Translational Neuroengineering Laboratory .
I am working on the synthesis of small molecule heparin glycomimetics to discover the structure-activity relationship in terms of growth factor binding and how this binding will modulate the peripheral nerve regeneration in vitro and in vivo models.
Vincenzo Scamarcio, Ph.D. student
I am a Ph.D. student at SuNMIL and Computational Robot Design & Fabrication Lab (CREATE-LAB) .
My research tackles the challenge of introducing flexible automation to streamline standard laboratory practices while understanding its current limitations. We made significant headway with a two-armed mobile robotic platform, by automating the labor-intensive protocol of determining micelles’ CMC by interfacing it with various instruments. Within this project, we also built simpler robots to investigate whether they can offer flexibility. For instance, we deployed Cartesian robots for tasks like bi-manual manipulation, handling tissue explants, and evaluating bacterial plates for antibiotic resistance using pattern recognition through computer vision.
Currently, we are extending the capabilities of the mobile robotic platform to execute various autonomous protocols, integrating machine learning and AI to allow repeated experiments without human input. We also test the system’s adaptability in physically reconfigured lab spaces, ensuring consistent interaction with labware through computer vision and tactile feedback.
As lab automation becomes increasingly prevalent in high-throughput experiments due to higher reproducibility and efficiency, the need for affordable, reconfigurable systems grows for smaller labs. Our future directions include exploring how mobile robots can ease the barrier to flexible automation and investigating modular robotic systems with interchangeable parts and programming capabilities that can be quickly reconfigured.
Ding Ren, Ph.D. student
In this project, I aim to discover novel protein cancer biomarkers. I extract lipoprotein nanoparticles from human serum/plasma, and use mass spec to analyze their proteomics. Preliminary results has shown that lipoprotein nanoparticles include far more information than serum on cancer detection, and 100+ proteins can be distinguished as significantly up-or-down regulated. This method can also separate non-metastatic cancer from metastatic ones, estimate prognosis, and help evaluate treatment effectiveness. In the future, I want to dig deeper into the structural analysis of lipoprotein nanoparticles, and use more advanced approach to analyze the data. Specific fingerprints are expected for each cancer, which may help to diagnose it more precisely.