Antibiotics have been widely used over the last century. Their modes of action have been extensively studied, mostly using model bacteria and common pathogens. As we move away from these model organisms, the way antibiotics affect diverse bacterial species or strains can be unexpected compared to what is described in textbooks. Our objective is to understand the molecular mechanism(s) that underline(s) the phenotypical heterogeneity that we observe during/after antibiotic treatment of diverse bacterial strains and species.
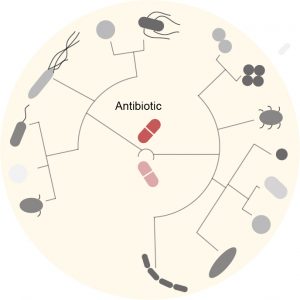
Antibiotic resistance is one of the major health challenges of our time. The overuse of antibiotics combined to the rapid adaptation power of bacteria has led to the emergence of bacteria that resist every single antibiotic available on the market. Bacteria can become resistant to antibiotics through genetic mutations or by taking up exogenous DNA from resistant surrounding bacteria, a phenomenon called horizontal gene transfer. Although we are currently witnessing a rapid increase and spread of antibiotic resistance elements worldwide, we know very little about the molecular mechanisms influencing antibiotic resistance in natural communities. Our objective is to better understand how antibiotic resistance emerges and spreads between human gut microbes.
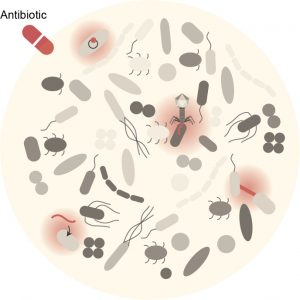
Antibiotic treatment is associated with two major health problems: the selection for antibiotic resistance and the collateral damage on the gut microbiota, which has recently been associated with short- and long-term health problems. There is therefore an urgent need to identify new molecules or new strategies to inhibit (resistant) bacteria. Here, our objective is to develop novel approaches to target pathogens while protecting our gut microbiota.