Spatial proteomics, awarded as Method of the year 2024 by Nature¹, offers unique insights into the distribution of proteins within tissues, which is crucial for a deeper understanding of complex biological processes such as cellular and tissue development, disease progression mechanisms, and biomarker discovery.
Our facility serves as a dedicated partner in advancing research excellence through cutting-edge technology, specialized expertise, and personalized support. Commited to our mission, we empower researchers to achieve their scientific goals while continuously bringing innovative technologies to our community.
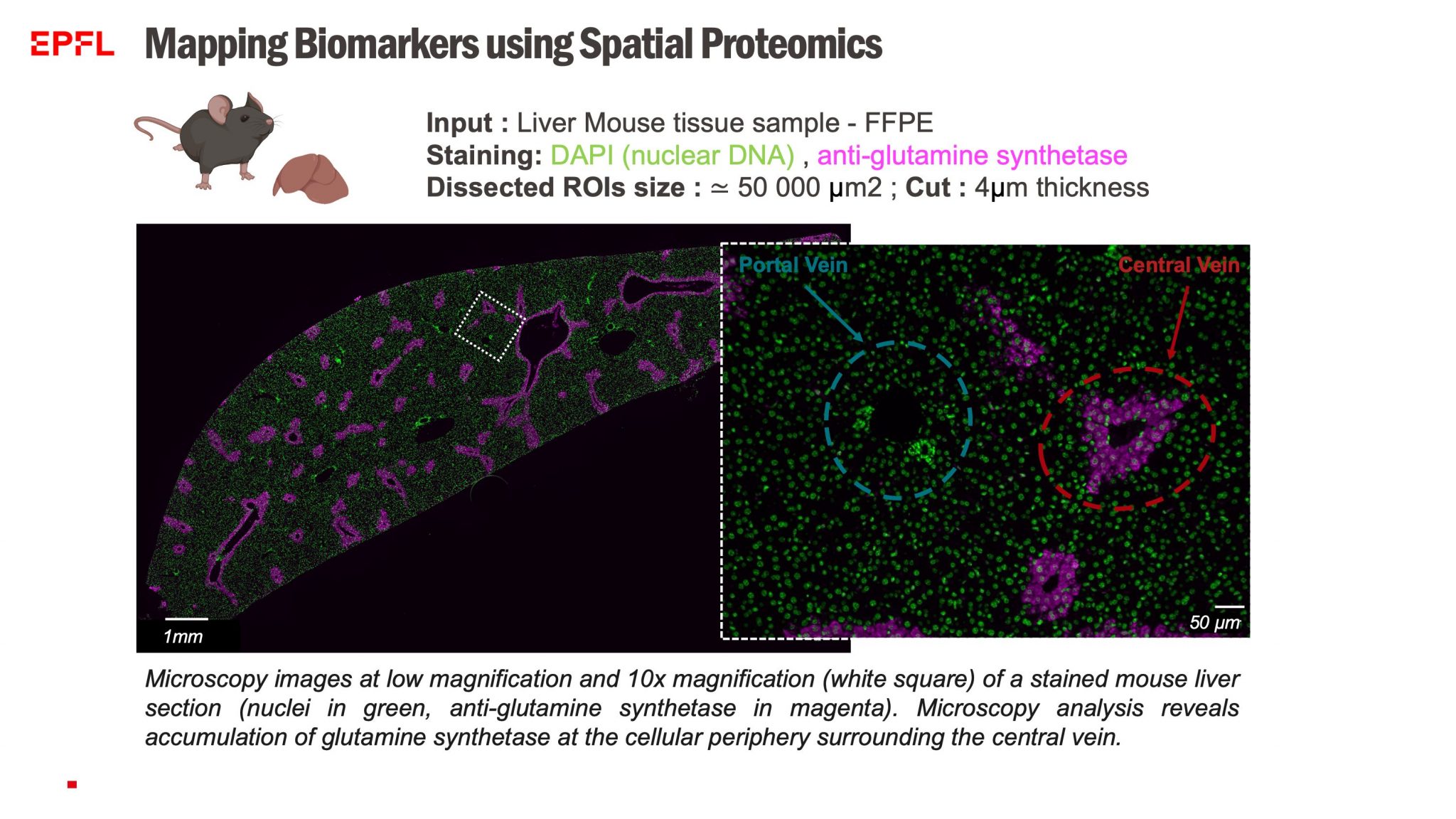
In 2024, PCF partnered with PTH and BIOP to establish spatial microproteomics capabilities for both fresh and fixed tissues.
We proudly introduce our cutting-edge spatial proteomics service, representing our latest advancement in empowering scientific discovery.
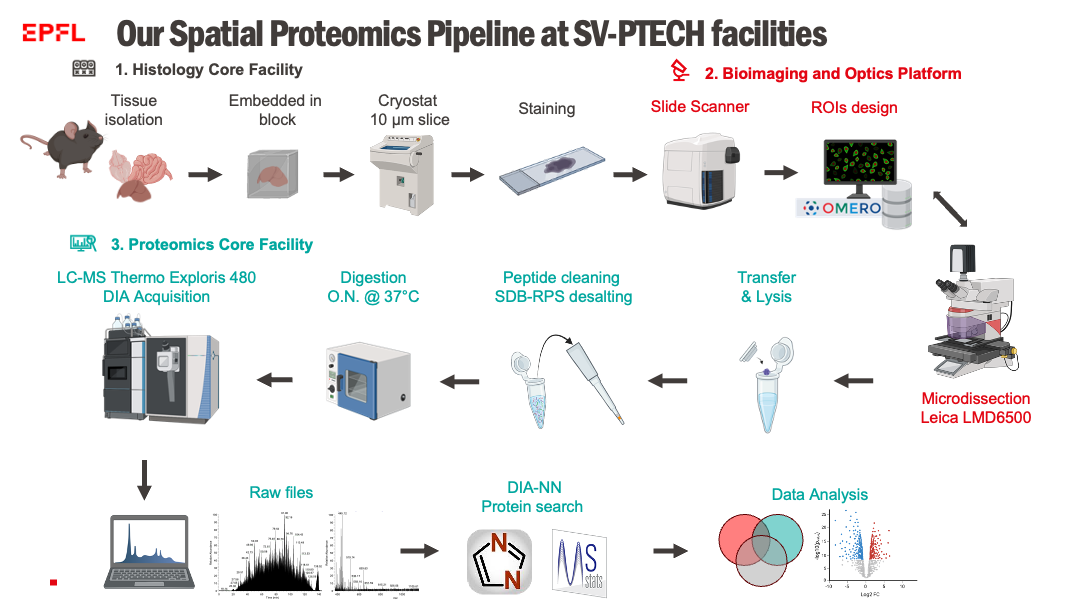
We have developed a generalizable spatial proteomics pipeline based on the DVP strategy (Deep Visual Proteomics²). Recognized for its impact³, our approach includes :
- Laser-Cut Tissue Microdissection – Allowing precise selection of regions of interest at the cellular level.
- Optimized Sample Preparation – Enabling extended protein characterization from as little as a few dozen cells.
- Bottom-Up Mass Spectrometry (MS) Analysis – Utilizing data-independent acquisition (DIA) methods specifically designed for low-input samples.
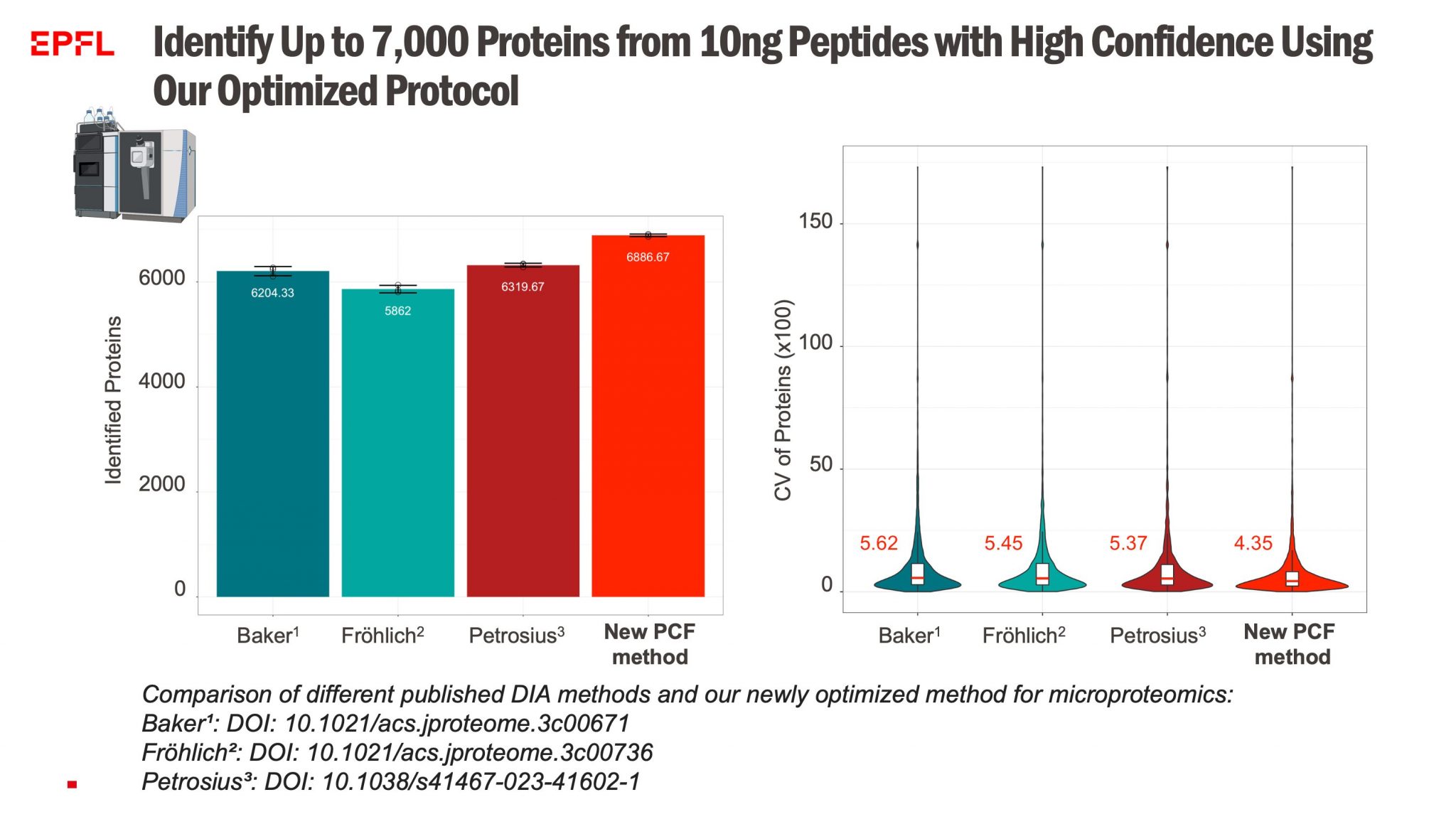
- Extended Protein Quantification – Effective and reliable quantification of up to 7,000 proteins from minimal starting sample quantities.
- Efficient Workflow – Optimized processes ensure timely data delivery while maintaining high-quality results.
- Versatility – Adaptable to various sample types, animal and human tissues, biopsies, organoids, and organs-on-chip.
- Fixation Compatibility – Supports various fixation methods, including FFPE blocks, fresh frozen, fixed frozen, and PFA/GA fixatives.
- Cost-Effective – An affordable analytical solution without compromising data quality.

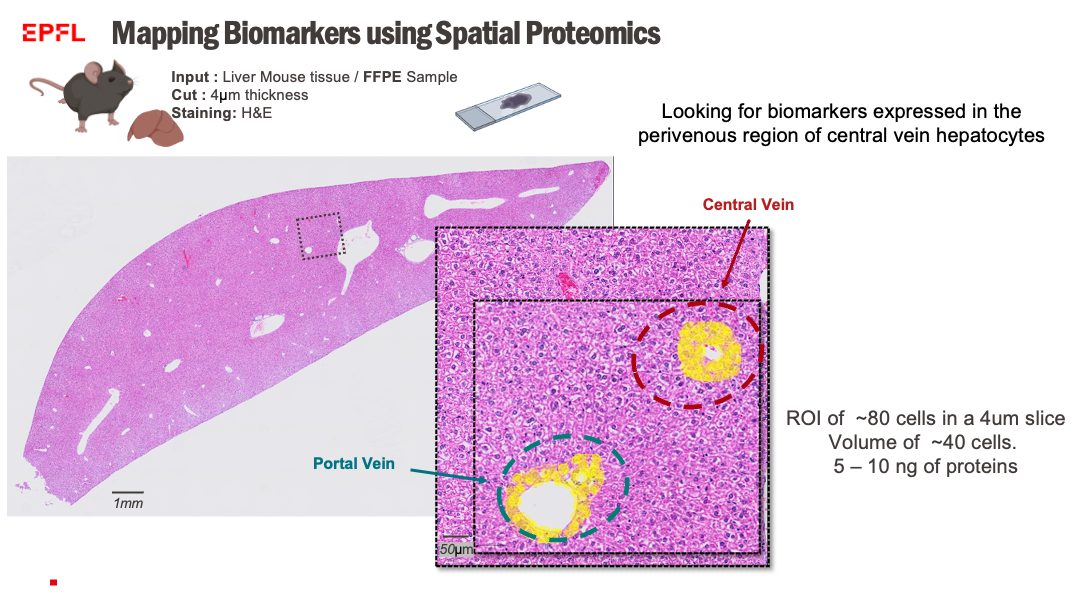
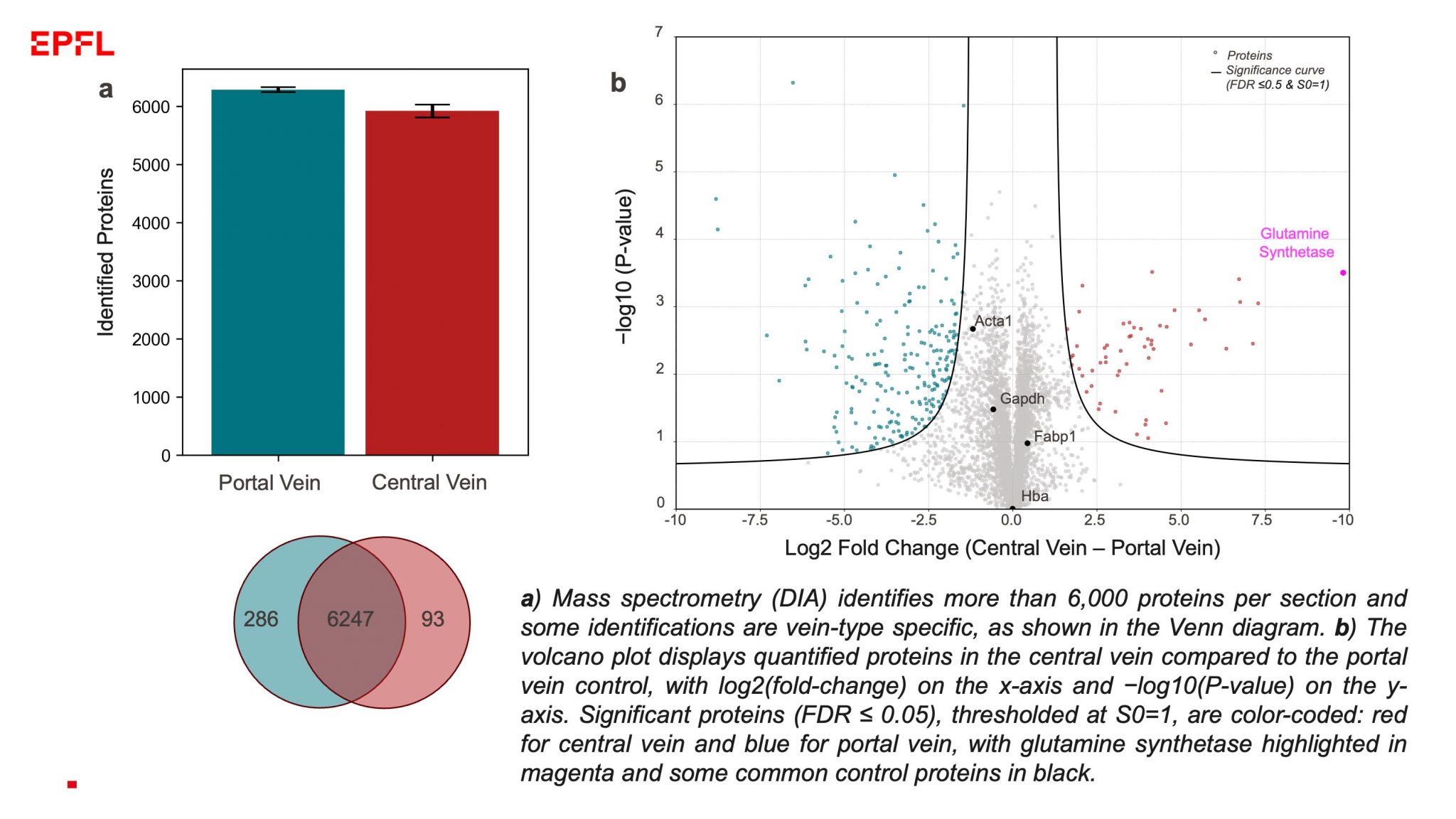
Researchers are invited to utilize our spatial proteomics pipeline for their projects. We offer:
- Consultation: Discuss your research goals and how our pipeline can assist
- Customized Workflow: Adapt the pipeline to suit your specific samples and objectives
- Expert Support: Guidance from our experienced team throughout the project
- Analysis Support: Assistance with data analysis and interpretation.
Interested in learning more or embarking in Spatial Proteomics?
Reach out!
- Email: Thibault Courtellemont (Project Leader Spatial Proteomics) ; Maria Pavlou (Head of PCF) ;
- Phone:+41 21 693 18 27
- Address: Plateformes Technologiques – SV, EPFL, Station 15, 1015 Lausanne, Switzerland
Literature:
1. Method of the Year 2024: spatial proteomics. Nat Methods 21, 2195–2196 (2024). https://doi.org/10.1038/s41592-024-02565-3
2. Mund, Andreas et al. “Deep Visual Proteomics defines single-cell identity and heterogeneity.” Nature biotechnology vol. 40,8 (2022): 1231-1240. doi:10.1038/s41587-022-01302-5
3. PCF development awarded as best oral presentation during LS2 Proteomics 2024

