Selected papers
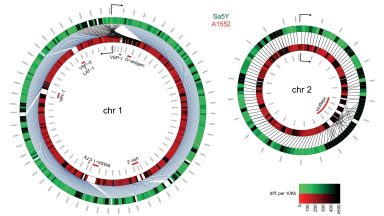
Vibrio Cholera genome analysis
We compared genomes of different strains, provided circular plots to visualize the differences, and use the strain differences to precisely detect transferred genomic regions from one ‘prey’ strain to one ‘predator’ strain.
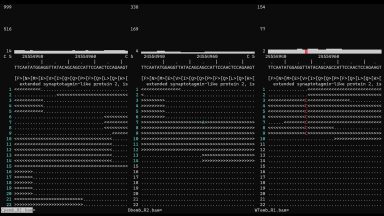
WGS, Exome, RNAseq analysis
We have the skills to analyze RNAseq data beyond the traditional differential gene expression analysis and identify SNPs, RNA editing, transcripts or splicing events present only in some samples, and highlight differences between samples at the nucleotide level.
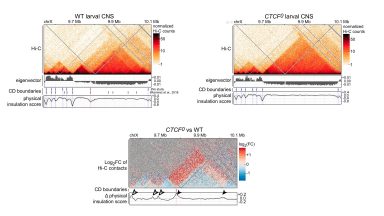
HiC
Processing of HiC samples to determine TAD boundaries, and detect chromosome contacts differing between strains
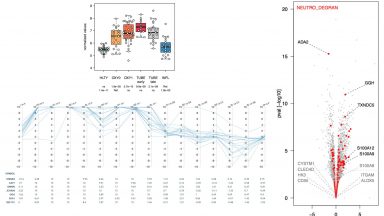
Transcriptomics
Mining transcriptomic Signature Differences Between SARS-CoV-2 and Influenza Virus Infected Patients